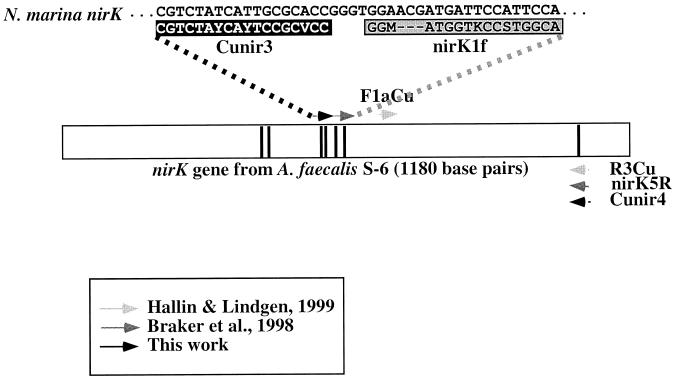
FIG. 1.
Locations of PCR primer pairs for the nirK gene relative to the nirK sequence of Alcaligenes faecalis S-6. Vertical bars indicate the locations of the seven amino acids involved in copper binding. Primers F1aCu and R3Cu (15) yield a 473-bp product, while nirK1F and nirK5R (5) and Cunir3 and Cunir4 (this study) yield 514- and 540-bp products, respectively. The sequence of the Cunir3 target site in the N. marina nirK gene was verified from the sequence of an additional 300-bp fragment which overlaps the 5′ end of the Cunir3-4 fragment (results not shown). Note that the target region for nirK1F spans a region with a 3-bp insertion (deletion) in the N. marina (A. faecalis) nirK sequence. Y = C or T; V = A, C, or G; M = A or C; K = G or T; S = G or C.