Figure 22.
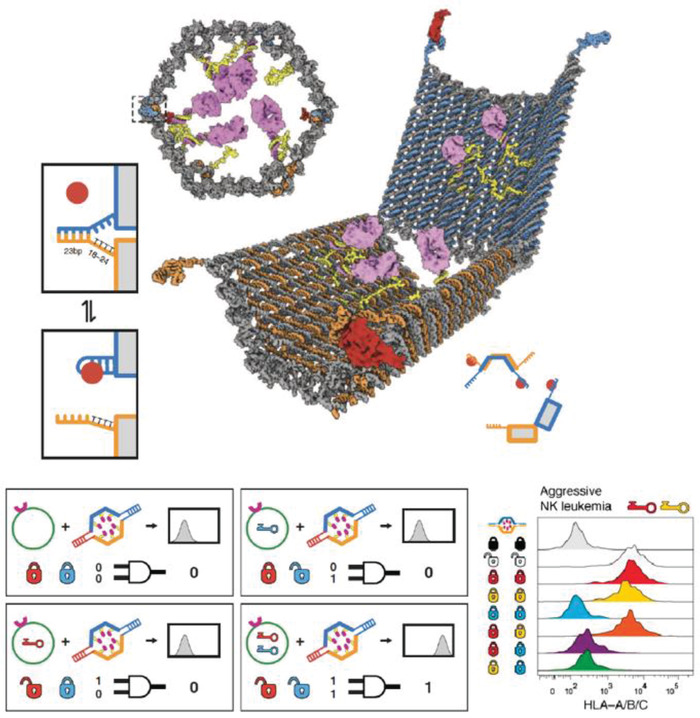
Reconfigurable DNA “nanorobot” used for controlled drug delivery. Top: design schematic of the DNA origami nanorobot in front, closed view and open view. Pink domains represent a protein payload. The aptamer lock mechanism (shown in two boxes on the left) is based on aptamer (blue) hybridization with a partially complementary strand (orange) in the closed state. In the presence of antigen “key” (shown as a red circle), the aptamer binds to the antigen, dehybridizes with the complementary strand, and consequently unlocks the nanorobot. Bottom: each nanorobot can be locked with two aptamer locks that are specific to different antigen keys. The two locks function as an AND logic gate where only in the presence of both antigen keys the nanorobot will unlock (indicated as signal 1). This was characterized by a histogram of fluorescence versus cell counts (right). Shown here are results of incubating a nanorobot loaded with fluorescent antibody fragments against human leukocyte antigen (HLA‐A/B/C) and with several different lock combinations based on three aptamers—41t, against platelet‐derived growth factor (PDGF), shown as a red lock; TE17, shown in yellow; and sgc8c, shown in blue. Black and white “locks” indicate controls of always closed and open nanorobots, respectively. Flow cytometry results (observing anti‐HLA‐A/B/C fluorescence) of incubation of the eight different nanorobots with aggressive NK leukemia cells that express antigens against PDG and TE17 aptamers. Reproduced with permission.[[qv: 48b]] Copyright 2012, American Association for the Advancement of Science.
