Figure 4.
Scc2 and Mediator physically interact
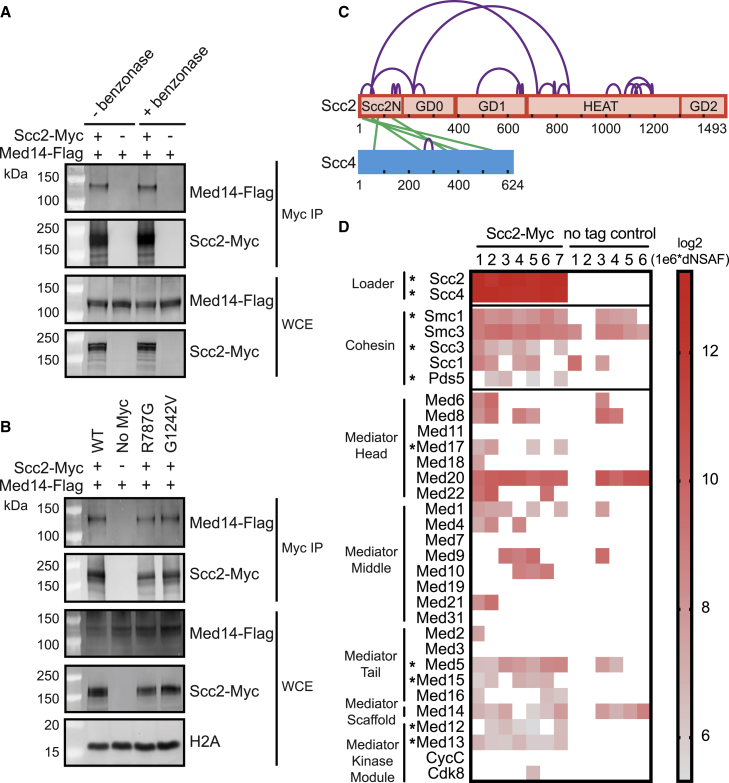
(A) Scc2 and Med14 interact by co-immunoprecipitation independent of DNA. Myc tagged Scc2 was immunoprecipitated from WCE. Co-precipitation of Flag-tagged Med14 was analyzed by immunoblotting. Benzonase treating WCE to remove DNA did not affect the observed co-precipitation.
(B) R787G and G1242V mutations do not affect the Scc2-Med14 interaction. Immunoprecipitations from the lysate of Myc tagged wild type, no tag, R787G, and G1242V Scc2 were analyzed for coIP of Med14-Flag by immunoblotting. The ratio of Scc2 to Med14 in the Scc2 mutants is comparable with WT in the Myc pulldowns.
(C) Diagram indicating inter- (green) and intra- (purple) protein cross-links of the Scc2-Scc4 complex. Positions of Scc2 domains are marked.
(D) Mediator is detected by mass spectrometry of purified Scc2. Cross-linking mass spectrometry was performed on Myc tagged Scc2 purified from WCE with anti-Myc magnetic beads with on bead disuccinimidyl sulfoxide (DSSO) cross-linking and on bead digestion. The heatmap of 1e6 log2 transformed dNSAF values shows enrichment of Mediator subunits in seven MS runs with purified Scc2-Myc compared with six no tag control runs. The loader and cohesin subunits are included as a positive control. Asterisks indicate statistical significance at p < 0.05.
See also Table S2 and Figures S4 and S5.