Figure 6.
Mediator facilitates Scc2 recruitment
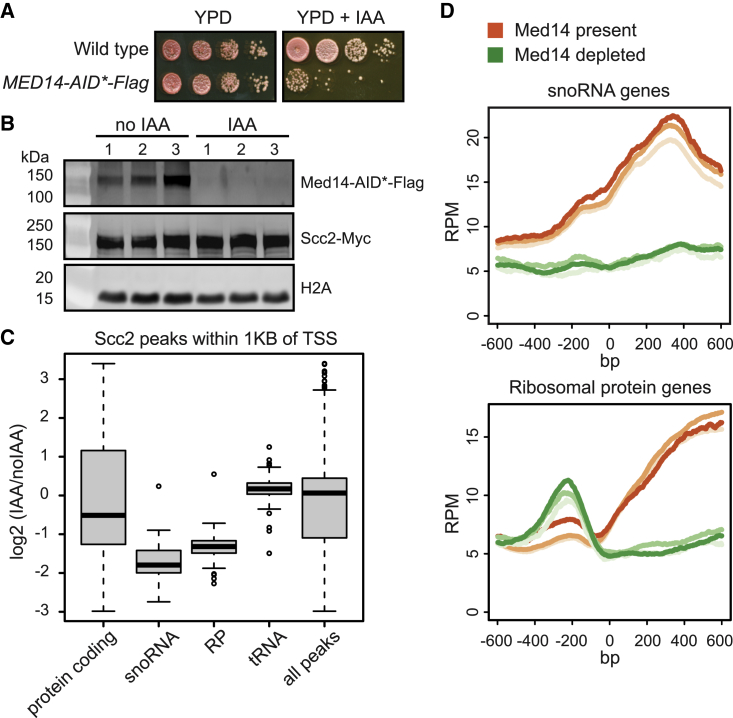
(A) Validation of Med14degron in Scc2-Myc tagged strain used for calibrated ChIP-seq. 10-fold serial dilutions were spotted on YPD and YPD+IAA.
(B) Med14degron is depleted in the presence of IAA. Cells grown in triplicate were synchronized in G1 with alpha-factor, Med14degron was depleted with IAA, and cells were released into nocodazole-imposed mitotic arrest. Cells were then cross-linked with formaldehyde. A portion of the culture was removed before cross-linking for gel electrophoresis and levels of Med14 and Scc2 were monitored by immunoblot. H2A served as a loading control.
(C) Scc2 peaks are most affected at snoRNA and RP genes after Med14 depletion. A spike-in of Mis4-Myc tagged S. pombe was added to each culture for calibration, and the Scc2-Myc immunopurified chromatin was extracted for ChIP-seq. The experiment was performed in triplicate with Med14 depleted (IAA) and Med14 present (noIAA). Peaks within 1 kb of transcription start sites were mapped to genes by biotype. The boxplot shows the log2 ratio of Scc2 peaks with Med14 depleted over no depletion (IAA/noIAA).
(D) Metagene analysis of the ChIP-seq data shows that Scc2 peaks decrease at snoRNA and ribosomal protein genes when Med14 is depleted.
See also Figure S6.