Abstract
Glucocorticoid stress hormones are powerful modulators of brain function and can affect mood and cognitive processes. The hippocampus is a prominent glucocorticoid target and expresses both the glucocorticoid receptor (GR: Nr3c1) and the mineralocorticoid receptor (MR: Nr3c2). These nuclear steroid receptors act as ligand‐dependent transcription factors. Transcriptional effects of glucocorticoids have often been deduced from bulk mRNA measurements or spatially informed individual gene expression. However, only sparse data exists allowing insights on glucocorticoid‐driven gene transcription at the cell type level. Here, we used publicly available single‐cell RNA sequencing data to assess the cell‐type specificity of GR and MR signaling in the adult mouse hippocampus. The data confirmed that Nr3c1 and Nr3c2 expression differs across neuronal and non‐neuronal cell populations. We analyzed co‐expression with sex hormones receptors, transcriptional coregulators, and receptors for neurotransmitters and neuropeptides. Our results provide insights in the cellular basis of previous bulk mRNA results and allow the formulation of more defined hypotheses on the effects of glucocorticoids on hippocampal function.
Keywords: corticosteroid receptors, hippocampus, single‐cell RNA sequencing, stress hormones, transcription regulation
1. INTRODUCTION
In the brain, stress responses and memory formation are essential to cope with changes in the environment. 1 The hippocampus is crucial in these processes, and highly sensitive to fluctuating levels of glucocorticoid (GC) stress hormones. 2 , 3 GC levels naturally vary along the day following circadian and ultradian rhythms, 4 and basal levels of endogenous GCs in the hippocampus are necessary for neuronal integrity, growth, differentiation and synaptic plasticity. 5 Although acute stress induces only a temporary deviation from this balance, chronic stress or excessive GC exposure can threat the hippocampal homeostasis. All of these effects are mediated by the two types of corticosteroid receptors that are expressed in the brain: the glucocorticoid receptor (GR) and the mineralocorticoid receptor (MR). GR and MR are nuclear steroid receptors that can act as ligand‐dependent transcription factors (TFs). MR has a high GC affinity (K d ~ 0.5 nm) and accordingly is activated substantially at basal hormone levels. GR has a lower affinity (K d ~ 5 nm) and is therefore responsive to circadian GC peaks and fluctuations in the stress range. 6 Binding studies, immunohistochemistry and in situ hybridization showed that expression of the Nr3c2 gene (coding for MR) is mainly restricted to the limbic brain, specifically the hippocampus, whereas the Nr3c1 gene (coding for GR) is widely expressed throughout the brain. 7
To date, all genome‐wide studies on GR‐ and MR‐mediated transcription in the hippocampus have been conducted with bulk tissue mRNA measurements. However, the hippocampus is a complex brain structure with a wide diversity of neuronal as well as non‐neuronal cells, and with a particular spatial organization. Single‐cell RNA sequencing (scRNA‐seq) has allowed for a large‐scale comprehensive molecular classification of cell types in the brain. 8 , 9 , 10 The Allen Institute for Brain Science recently sequenced approximately 1.2 million cells covering all regions of the adult mouse isocortex and hippocampal formation, identifying almost 380 subtypes of cells. The hippocampal data includes information on glutamatergic neurons from the dentate gyrus (DG) and cornu ammonis regions, GABAergic neurons, astrocytes, oligodendrocytes, microglia and endothelial cells. 11 Our previous in situ hybridization‐based analysis on whole brain revealed spatially specific co‐expression patterns of Nr3c1 and Nr3c2 with genes that are responsive to GCs or involved in nuclear receptor transcriptional regulation. This suggested mechanisms for regional and cellular functional specificity of GC signaling. 12 The advances in scRNA‐seq carry with them new computational methods to address such co‐expression at the cell type level, and allow the reconstruction of TF downstream pathways. 13 , 14 , 15
In the present study, we used existing scRNA‐seq data 11 to molecularly characterize the cellular heterogeneity of GR and MR signaling in the adult mouse hippocampus. We assessed cell type expression specificity of GR and MR downstream target genes to identify putative markers for GC responsiveness in particular cell types. Furthermore, we looked into GR and MR co‐expression with sex hormone receptors, transcriptional coregulators, and receptors for neurotransmitters and neuropeptides to define for each cell type the potential pathways that may interact with hippocampal GC signaling.
2. MATERIALS AND METHODS
2.1. scRNA‐seq data resources
The present study is based on the 10x scRNA‐seq dataset published by the Allen Institute for Brain Science 11 (https://portal.brain‐map.org/atlases‐and‐data/RNA‐seq/mouse‐whole‐cortex‐and‐hippocampus‐10x). Briefly, the single cells were isolated from 16 different regions of the isocortex and the hippocampal formation from 54 male and female mice. The Allen Mouse Brain Common Coordinate Framework version 3 (CCFv3) ontology was used to define brain regions for profiling and boundaries for dissections. scRNA‐seq data from the regions of interest were generated using 10x Genomics Chromium. For downstream processing, cells with <1500 detected genes as well as doublets were filtered out. The data was then clustered, and cluster names were assigned based on the Allen Institute proposal for cell type nomenclature (https://portal.brain‐map.org/explore/classes/nomenclature). The topology of the taxonomy allowed to define the sex of the mouse from which the cells were isolated, the regions of interest, cell classes (glutamatergic, GABAergic or non‐neuronal) and subclasses. 11 , 16 This information was stored in the metadata table.
2.2. scRNA‐seq data metrics and pre‐processing
The metadata was used to subset cells of the hippocampus region from the gene expression matrix. We selected for 13 subclasses of hippocampal cells. The final gene count matrix consisted of 77,001 cells for 26,139 genes (Figure 1) and was pre‐processed in R, version 3.6.1 (R Foundation for Statistical Computing) according to the Seurat, version 3.1.5 (https://satijalab.org/seurat) pipeline for quality control, normalization and analysis of scRNA‐seq data using the following criteria: min.cells = 1, min.features = 100, normalized.method = LogNormalize, scale.factor = 10000, selection.method = “vst”, nfeatures =2000. The gene counts were normalized and log‐transformed across all cells, which allowed for statistical comparison between cells and cell types, as described previously. 17 We performed principal component analysis and we selected the top 50 PCs as input for the t‐distributed stochastic neighbor embedding (t‐SNE) dimensional reduction. Finally, the transcriptomic data was analyzed and displayed using Seurat visualization tools (Figure 1).
FIGURE 1.
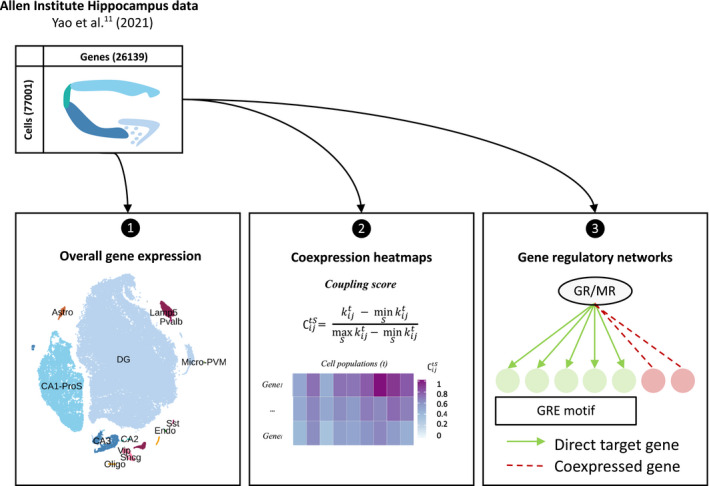
Schematic overview of the research strategy. Abbreviations: Astro, astrocytes; Oligo, oligodendrocytes; Endo, endothelial cells; micro‐PVM, microglia/perivascular macrophages; Lamp5, lysosomal associated membrane protein family member 5; Vip, vasoactive intestinal peptide; Pvalb, parvalbumin; Sncg, synuclein gamma; Sst, somatostatin; DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3; , coupling score, GR, glucocorticoid receptor; MR, mineralocorticoid receptor; GRE, glucocorticoid response element
2.3. Bulk RNA sequencing of mouse ventral hippocampus
The animal study was approved by the ethics committee of local Animal Committee of the University of Amsterdam. Eight‐week‐old C57BL/6 J male mice were group‐housed by four in conventional cages under a 12:12 hour light/dark photocycle and had access to food and water available ad libitum. Mice received an injection of either 3 mg.kg–1 corticosterone (n = 4) or vehicle (n = 4) between 9:00 and 10:00 a.m. Mice were killed by decapitation 3 h after injection. The ventral hippocampus was collected for mRNA sequencing (RNA‐seq). Total RNA was isolated with the NucleoSpin® RNA kit (Macherey‐Nagel) and RNA quality was assessed using the RNA 6000 Nano kit on Bioanalyzer (Agilent). All samples had an RNA Integrity Number over 6.5 with a 28/18s ratio over 1, and therefore were considered suitable for sequencing. Aliquots of total RNA samples were sent for transcriptome sequencing at BGI Genomics. Stranded mRNA libraries were constructed and 100‐bp paired end sequencing was performed on the DNBseq platform, resulting in over 20 million reads per sample. RNA‐seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE184924. The Gentrap pipeline, published as part of Bio Pipeline Execution Toolkit (Biopet, https://biopet‐docs.readthedocs.io), was used for reads quality control, alignment and quantification. Quality control was performed using FastQC and MultiQC. Reads were aligned 10 mm using GSnap aligner, version 2017‐09‐11. The gene‐read quantification was performed using HTSeq‐count, version 0.6.0. HTSeq‐count output files were merged into a count matrix as input for differential gene expression analysis. DEseq2, version 1.29.4, 18 was used for normalization of the data (median of ratio’s method) and identification of differentially expressed genes in R, version 3.4. The differential expression analysis, resulting in 16,839 genes in the analysis. The contrast between vehicle and corticosterone groups was analyzed for differential expression in a pairwise comparison. An false discovery rate adjusted p value of .05 was used as a cut‐off to determine differentially expressed genes.
2.4. Selection of gene sets
Steroid receptors: This gene set contains the stress and sex hormones nuclear steroid receptors, the GR (Nr3c1 – nuclear receptor subfamily 3 group C member 1), the MR (Nr3c2 – nuclear receptor subfamily 3 group C member 2), the androgen receptor (Ar), the progesterone receptor (Pgr), and the estrogen receptors α and β (Esr1 and Esr2).
GR and MR target genes: This set of genes is based on previous transcriptomic studies in rodent brain and neuronal cells after glucocorticoid treatment, 19 our recent RNA‐seq results in mouse ventral hippocampus after corticosterone injection, and two chromatin immunoprecipitation followed by sequencing (ChIP‐seq) studies on GR and MR after injection with either 0.3 or 3 mg.kg–1 corticosterone in rats. 20 , 21 The criteria for ‘target genes’ were (1) regulation by GCs in previously published studies on rodent brain and (2) in our recent transcriptomic results, given that these exclusively represent mouse hippocampus; (3) the direction of regulation had to be consistent in all reporting studies; and (4) the gene had to be associated with a binding site for either GR, MR or both receptors according to the two ChIP‐seq studies that we used. The latter were in rat hippocampus, but it has become apparent that functional GC response elements (GREs) tend to be evolutionary conserved. 22 , 23
Coregulators: The gene set of GR and MR AF‐2 coregulators was based on previous profiling analysis published by Broekema et al. 24
Neurotransmitter and neuropeptides receptor repertoire: We aimed for an exhaustive list of genes for the adrenergic, serotoninergic, cholinergic and dopaminergic receptors according to the HUGO Gene Nomenclature Committee at the European Bioinformatics Institute (HGNC database: https://www.genenames.org). The neuropeptides receptors list was based on the HGNC database and the previous study from Smith et al. 25 on intracortical neuropeptide networks.
2.5. scRNA‐seq coupling matrices for Nr3c1 and Nr3c2 co‐expression profiles
A coupling score of Nr3c1 and Nr3c2 with genes of interest was calculated to rank their co‐expression. First, we calculated the average expression of each gene of interest i in cell type t (), where t is one of the 13 cell types in the adult mouse hippocampus. For each corticosteroid receptor (Nr3c1 and Nr3c2), we calculated the coupling score as previously described, 25 as , where and is one of the gene sets described earlier, and . For each gene set , we calculated the normalized coupling score (Figure 1):
2.6. pySCENIC: Assessment of GR and MR single cell gene regulatory network activity
The gene expression matrix of the clustered hippocampus scRNA‐seq dataset underwent the scalable Python SCENIC (pySCENIC) (https://pyscenic.readthedocs.io) workflow for single‐cell gene regulatory network (GRN) analysis as described by Van de Sande et al. 15 pySCENIC reconstructs GRNs (i.e., TFs together with their target genes) and assesses the de novo GRN activity in individual cells (Figure 1). The pySCENIC workflow, version 0.10.3, was performed under Python, version 3.8.5 (https://www.python.org) and the output was then processed with Seurat, version 3.1.5 in R, version 3.6.1.
2.7. Differential expression and GRN activity analysis of scRNA‐seq data
The gene count matrix for hippocampal gene expression and the GRN activity matrix underwent differential expression/activity analysis to identify genes specifically more expressed or GRNs specifically more active in certain cell types. Both differential analyses were performed using the Seurat FindAllMarkers function (Wilcoxon rank sum test) 17 in R, version 3.6.1. Furthermore, significant differences in gene expression throughout cell types or within one cell type were tested with a paired two‐sided Wilcoxon test (wilcox.test function) on average expression in R, version 3.6.1.
2.8. Code availability
Open‐source algorithms were used as described for single‐cell analysis methods 17 and GRNs analysis. 15 Details on how these algorithms were used, as well as the code for coupling score calculation, are available in the GitHub repository (https://github.com/eviho/10XHip2021_VihoEMG).
3. RESULTS
3.1. Nr3c1 (GR) and Nr3c2 (MR) expression show significant cell specificity across hippocampal cell types
Our approach aimed to describe the diversity of corticosteroid receptors Nr3c1 (GR) and Nr3c2 (MR) signaling networks in mouse hippocampal cell types, using publicly available scRNA‐seq data. We selected hippocampal cells from the Yao et al. 11 mouse brain dataset, which resulted in 77,001 cells, divided over 13 different cell types (Figure 2A). The most abundant cell types in this dataset were the DG and cornu ammonis 1/pro‐subiculum (CA1‐ProS) glutamatergic neurons with 58,566 and 13,221 cells, respectively. The two last glutamatergic neuron populations CA2 and CA3 contained 143 and 1899 cells, respectively. GABAergic neurons were divided into five subtypes containing between 49 and 1372 cells: parvalbumin (Pvalb), somatostatin (Sst), vasoactive intestinal peptide (Vip), synuclein gamma (Sncg) and lysosomal associated membrane protein family member 5 (Lamp5) positive neurons. Finally, the data revealed four non‐neuronal cell types: 488 astrocytes (Astro), 465 oligodendrocytes (Oligo), 73 endothelial cells (Endo) and 74 microglial cells/perivascular macrophages (micro‐PVM) (Figure 2B).
FIGURE 2.
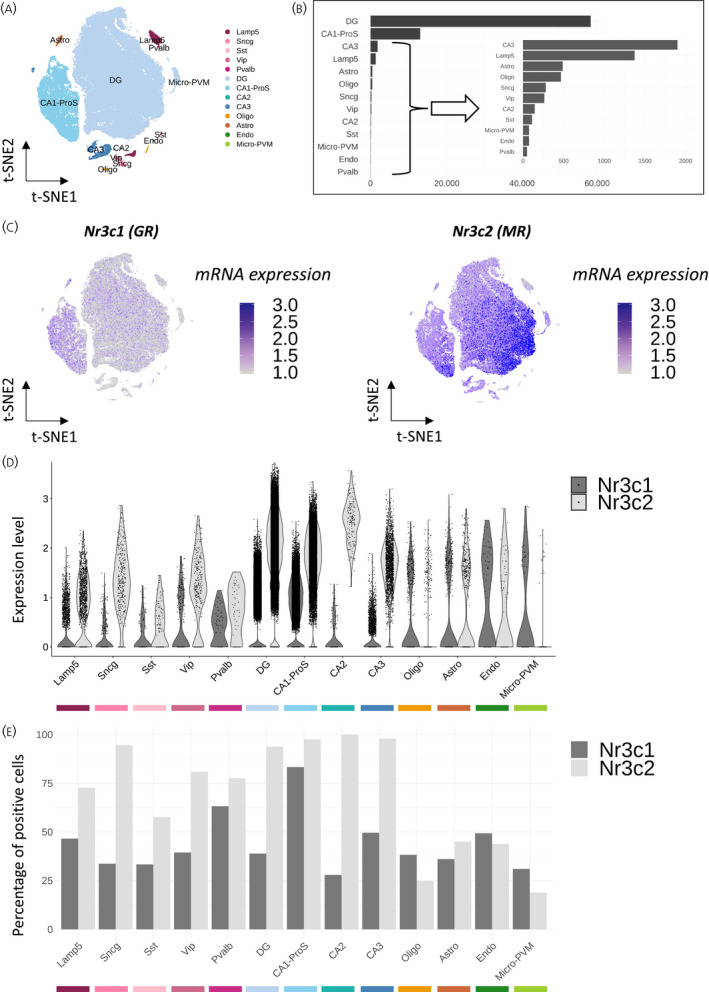
Cell type specificity of Nr3c1 and Nr3c2 expression in the adult mouse hippocampus. (A) Dimensional reduction (t‐SNE) representation of mouse hippocampal cells grouped by gene expression profile similarities and assigned to known cell types. (B) Number of cells per cell type within the dataset. (C) t‐SNE representation of Nr3c1 and Nr3c2 log‐normalized mRNA expression per cell, scaled from 1 to 3 (mRNA expression). (D) Violin plot of Nr3c1 and Nr3c2 log‐normalized expression (Expression level). (E) Bar plot of the percentage of cells positive for Nr3c1 and Nr3c2. Abbreviations: t‐SNE, t‐distributed stochastic neighbor embedding; Nr3c1, nuclear receptor subfamily 3 group C member 1; Nr3c2, nuclear receptor subfamily 3 group C member 2; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; Astro, astrocytes; Oligo, oligodendrocytes; Endo, endothelial cells; Micro‐PVM, microglia/perivascular macrophages; Lamp5, lysosomal associated membrane protein family member 5; Vip, vasoactive intestinal peptide; Pvalb, parvalbumin; Sncg, synuclein gamma; Sst, somatostatin; DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3
We assessed Nr3c1 and Nr3c2 relative expression levels throughout the hippocampal cell types. Although the t‐SNE representation clearly showed a significant higher expression level of Nr3c2 compared to Nr3c1 in the mouse hippocampus (log2FC = 2.82, p = .02) (Figure 2C), the data was biased towards the most abundant cell types (DG and CA1‐ProS). Per population, we observed a relatively higher expression of Nr3c2 compared to Nr3c1 in glutamatergic neurons, which was more pronounced in CA2 (log2FC = 3.74, p < .001) (Figure 2D). Nr3c2 was actually enriched in CA2 (log2FC = 0.53, p < .001) and the DG (log2FC = 0.32, p < .001) compared to other cell types (see Table S1). Interestingly, Nr3c2 was also more expressed than Nr3c1 in GABAergic neurons with the biggest difference in Sncg neurons (log2FC = 2.75, p < .001) (see Table S1). Nr3c1 was more expressed in non‐neuronal cell types with the biggest contrast in micro‐PVM cells where Nr3c2 was almost absent (Figure 2D). These differences in expression levels were in line with the percentage of cells expressing Nr3c1 and Nr3c2. Between 50% and 100% of neurons (glutamatergic and GABAergic) were positive for Nr3c2, whereas only CA1‐ProS, CA3 and Pvalb types passed the 50% threshold of positive cells for Nr3c1. Regarding non‐neuronal types, they contained <50% cells positive for either Nr3c1 or Nr3c2, with a slightly higher percentage of positive cells for Nr3c1 compared to Nr3c2 in oligodendrocytes, microglial and endothelial cells (Figure 2E).
Altogether, the results suggest a relatively higher basal expression of Nr3c2 in mouse hippocampal neurons and astrocytes, whereas Nr3c1 is relatively more expressed in oligodendrocytes, microglia and endothelial cells.
3.2. Classic GR and MR target genes differentially express across hippocampal cell types
Transcription‐dependent GC responsiveness of the hippocampus relies by definition on the presence of various GR and MR target genes. We investigated the basal expression of GC regulated genes in different hippocampal cell types. A limited class of genes is commonly measured in bulk brain mRNA to assess GC effects. 26 , 27 , 28 , 29 , 30 This set includes FK506‐binding protein 5 (Fkbp5), glucocorticoid‐induced leucine zipper protein (Tsc22d3), period circadian regulator 1 (Per1) and serum/glucocorticoid regulated kinase 1 (Sgk1). However, the scRNA‐seq data showed a clear heterogeneity for the basal expression of these genes in different hippocampal cell types (Figure 3A). Fkbp5 expression was predominant in glutamatergic neurons, particularly in the DG. In comparison, Tsc22d3 was more expressed in GABAergic neurons and non‐neuronal cells than Fkbp5. Furthermore, the basal expression of Per1 suggested high cell specificity, with high expression in only five neuronal cell types. Finally, Sgk1 was expressed in most hippocampal cell types, but was absent in astrocytes and endothelial cells (Figure 3A). The average expression was in line with the percentage of cells expressing the genes of interest. On average, 50% of glutamatergic neurons expressed Fkbp5, whereas 50% of GABAergic neurons expressed Tsc22d3. Sgk1 was more present in oligodendrocytes and microglia, whereas Tsc22d3 was more present in astrocytes and endothelial cells (see Figure S1A). Per1 was generally less expressed than any other classic target genes in the whole hippocampus, which might be partially explained by circadian variation (Figure 3A; see also Figure S1A). Although the analysis is performed on hippocampal basal gene expression, the results suggest an heterogenous and cell type‐specific response to GC signaling activation.
FIGURE 3.
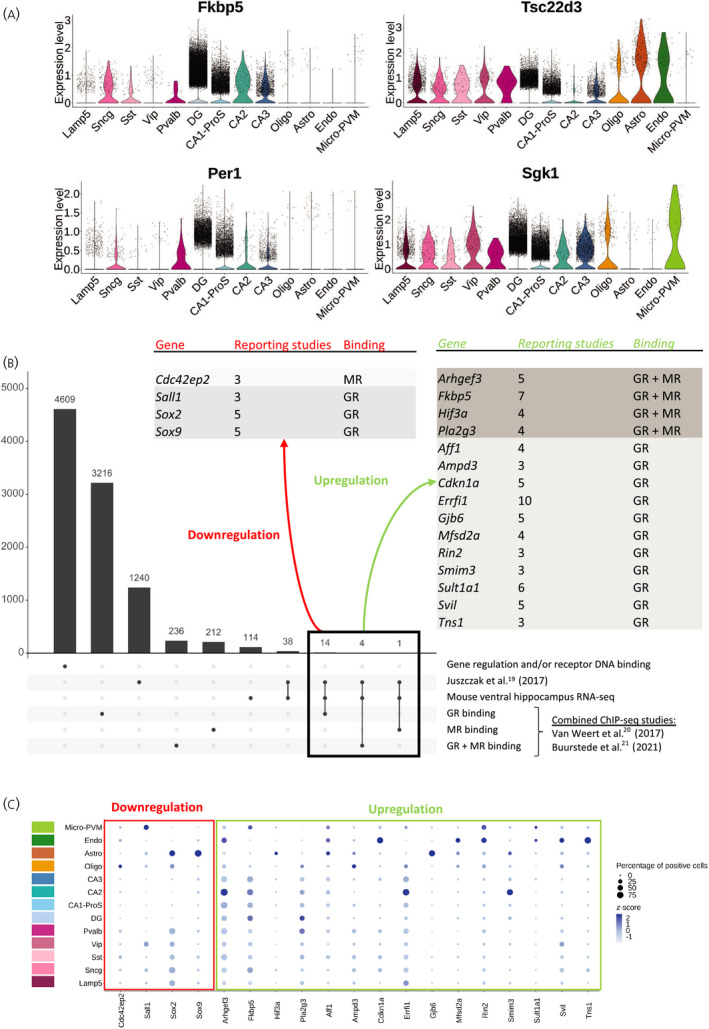
Cell type specificity of glucocorticoid target genes in the adult mouse hippocampus. (A) Violin plots representing the log‐normalized expression of commonly measured glucocorticoid responsive genes Fkbp5, Tsc22d3, Per1 and Sgk1 (Expression level). (B) List of new GR and MR target genes selection based on transcriptomic and DNA binding studies, associated with the number of transcriptomic studies reporting the gene (reporting studies), and DNA binding by GR, MR or both receptors (binding). (C) Dot plot representing both the centered log‐normalized average expression (z‐score) and the percentage of positive cells for the genes newly identified as GR and MR targets. Abbreviations: GR, glucocorticoid receptor; MR, mineralocorticoid receptor; ChIP, chromatin immunoprecipitation; RNA‐seq, RNA sequencing; Astro, astrocytes; Oligo, oligodendrocytes; Endo, endothelial cells; micro‐PVM, microglia/perivascular macrophages; Lamp5, lsosomal associated membrane protein family member 5; Vip, vasoactive intestinal peptide; Pvalb, parvalbumin; Sncg, synuclein gamma; Sst, somatostatin; DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3
Regarding MR‐specific target genes, MR binding to DNA on GREs was described to be associated with NeuroD factor binding 31 and Jdp2 was found as an MR target gene in conjunction with MR/NeuroD binding. At the basal level in the scRNA‐seq data, Neurod2 was mostly expressed in glutamatergic neurons and, although relatively few cells were positive for Jdp2, those expressing it were also glutamatergic neurons (see Figure S1B). Nr3c2 expression in the DG differed throughout the cell population (Figure 2C). Therefore, we assessed DG cells using a deeper level of clustering. DG cells could be further divided into six distinct subclusters. 11 , 16 The most abundant cluster was 125_DG, where both Nr3c2 and Neurod2 still showed different levels of expression across the cell cluster, with a similar overall pattern (see Figure S1C). This suggests that, despite differentially expressing Nr3c2 and Neurod2, cells in cluster 125_DG were not sufficiently divergent in the rest of their gene expression profile to be subdivided into more cell clusters. Jdp2 was mainly expressed in cluster 122_DG and 125_DG. However, in the absence of GC treatment, Jdp2 expression did not strongly correlate with the contrasted expression of Nr3c2 or Neurod2 in the DG (see Figure S1C).
3.3. A wider set of GC target genes further reveals GR and MR signaling heterogeneity across cell types
Although classic GC responsive genes already showed cellular heterogeneity of gene expression, we expanded the list of GC responsive genes to give a better recapitulation of cellular specificity of GR and MR signaling in the mouse hippocampus. We combined a published meta‐analysis on GC responsive genes in rodent and human brain (17 studies) 19 with a recent RNA‐seq dataset that we obtained in mouse ventral hippocampus, as well as ChIP‐seq data assessing GR and MR DNA binding in rat hippocampus 20 , 21 (see Table S2). This resulted in a list of 4609 genes either responsive to GC treatment or associated with a receptor binding site. Among those genes, 3216 reported GR‐specific binding to the DNA, 212 MR‐specific binding, and 236 reported both GR and MR binding. A total of 1240 genes were reported to be regulated in the previously published meta‐analysis, and 114 genes were GC responsive in our recent mouse hippocampus RNA‐seq dataset. We first selected for genes that were reported consistently in between the previously published meta‐analysis 19 and our transcriptomic analysis. This subset of 38 genes was further filtered for genes that reported DNA binding of either GR, MR or both receptors in the ChIP‐seq studies. In total, 19 genes survived all criteria and were reported in at least three transcriptomic studies. Of these, four genes were consistently downregulated and 15 were consistently upregulated. Cdc42ep2 was the only gene associated with MR binding, and a total of 14 genes were associated with exclusive GR binding and four genes were associated with both GR and MR binding, including Fkbp5 (Figure 3B). Tsc22d3, Per1 and Sgk1 were previously reported in both transcriptomic and ChIP‐seq studies but absent in the recent mouse hippocampus RNA‐seq dataset (see Table S2).
The new subset of GR and MR target genes was further analyzed in the hippocampus scRNA‐seq data. Similar to the classic GC responsive genes, the new targets displayed a large heterogeneity in cell type basal expression (Figure 3C). Genes known to be downregulated after GC treatment showed high specificity for non‐neuronal cell types. Cdc42ep2 was relatively more expressed in oligodendrocytes, Sall1 in microglia, and Sox2 and Sox9 in astrocytes. Among genes known to be upregulated after GC treatment, more than half were relatively more expressed in non‐neuronal cells in these basal conditions. However, Fkbp5 and Pla2g3 were predominantly neuron specific. Moreover, Arhgef3, Errfi1 and Smim3 were preferentially expressed in CA2 (Figure 3C). We also investigated the cell type specificity of genes known to be regulated by GCs but not associated with a receptor binding site. In this list of 19 genes, three were not detectable in the scRNA‐seq data (1810011O10Rik, Rhou, Lcn2). Many genes were highly expressed in astrocytes (e.g., Dio2), two downregulated genes (Abi3, Ccr5) were microglia specific, and three genes were widely expressed in neurons but at low levels, except for Ccng1 which was highly expressed and abundant in CA1‐ProS (see Figure S1D).
The results for GR and MR downstream target genes again highlighted the expression heterogeneity of GC target genes in mouse hippocampal cell types. Furthermore, under basal conditions, many target genes were specifically expressed in non‐neuronal cells. This indicates that transcripts from non‐neuronal cells may represent a substantial part of GC target genes.
3.4. Nr3c1 and Nr3c2 co‐expression with sex hormone receptors suggests cell type‐specific crosstalk
Corticosteroid receptors belong to the nuclear receptor superfamily that also includes the sex hormone receptors: the progesterone receptor (PR, coded by Pgr), androgen receptor (AR, coded by Ar), and estrogen receptors α and β (ERα and ERβ, coded by Esr1 and Esr2). Sex steroid receptors may interact with MR and GR, but direct interactions would by definition depend on presence and co‐expression. 32 , 33 , 34
Ar, Pgr, Esr1 and Esr2 were similarly expressed in cells that came from male or female mice in the scRNA‐seq with two subtle differences. Pvalb GABAergic neurons showed lower expression of Ar and Pgr in male cells, and CA3 had more positive cells and a slightly higher expression of Pgr in males. Esr1 and Esr2 were expressed in very few cells, with the highest expressing cell types being the DG granule cells and CA1‐ProS (Figure 4A).
FIGURE 4.
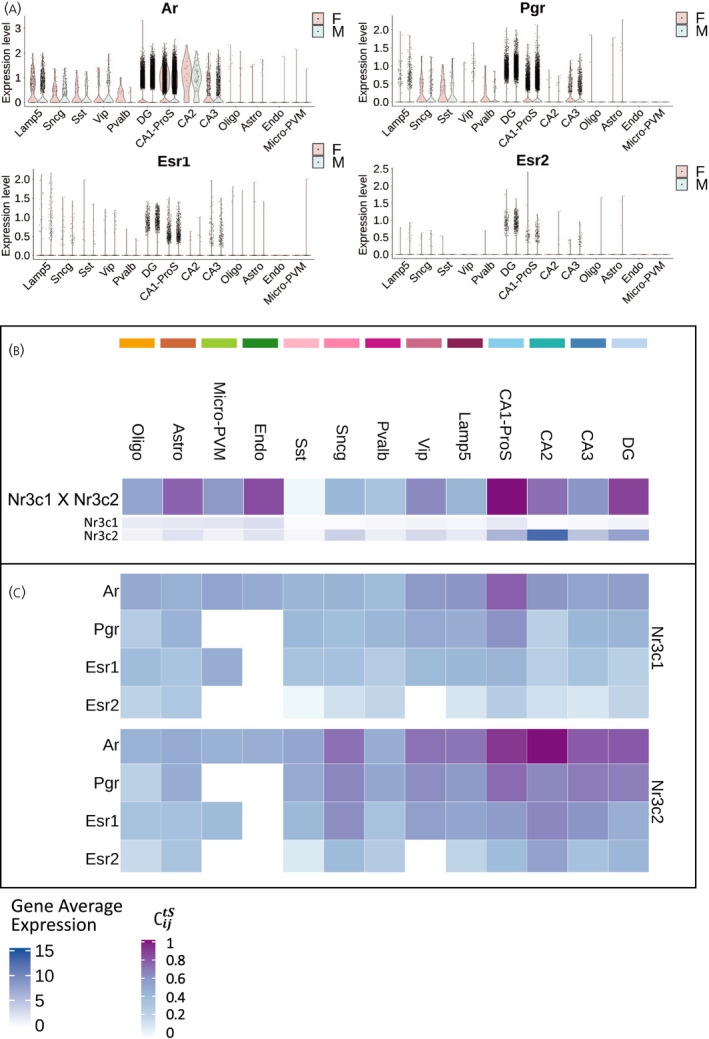
Cell type specificity of Nr3c1 and Nr3c2 co‐expression with sex hormone receptors. (A) Violin plots representing the log‐normalized expression (Expression level) of sex hormone receptors Ar, Pgr, Esr1 and Esr2 in cells obtained from female (F) and male (M) mice. (B) Heatmap representing the coupling score of Nr3c1 with Nr3c2, and their respective log‐normalized average expression in mouse hippocampal cell types (Gene Avg. Exp). (C) Heatmap representing the coupling score of Nr3c1 and Nr3c2 with sex hormone receptors Ar, Pgr, Esr1 and Esr2 in mouse hippocampal cell types. Abbreviations: Nr3c1, nuclear receptor subfamily 3 group C member 1 (glucocorticoid receptor); Nr3c2, nuclear receptor subfamily 3 group C member 2 (mineralocorticoid receptor); Ar, androgen receptor; Pgr, progesterone receptor; Esr1 and Esr2, estrogen receptors α and β; F, female; M, male; Astro, astrocytes; Oligo, oligodendrocytes; Endo, endothelial cells; micro‐PVM, microglia/perivascular macrophages; Lamp5, lysosomal associated membrane protein family member 5; Vip, vasoactive intestinal peptide; Pvalb, parvalbumin; Sncg, synuclein gamma; Sst, somatostatin; DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3
We next determined cell type‐specific co‐expression between stress and sex hormone receptors. For this, we calculated a coupling score based on basal average expression of pairs of genes in the different hippocampal cell types. Corticosteroid receptors (Nr3c1 and Nr3c2) showed the highest coupling score in CA1‐ProS and were also highly co‐expressed in the DG, CA2, endothelial cells and astrocytes (Figure 4B; see also Table S3). The highest coupling score between stress and sex hormone receptors was found in neuronal cells. Nr3c1 particularly co‐expressed with Ar and Pgr in CA1‐ProS, whereas Nr3c2 not only co‐expressed with Ar mainly in glutamatergic, Lamp5, Vip and Sncg neurons, but also with Pgr in CA1‐ProS (Figure 4C; see also Table S3). The coupling scores between Nr3c1 and Nr3c2 and estrogen receptors were very low because of the absence of Esr1 or Esr2 expression in most cells. The highest coupling score for Esr1 and Nr3c2 was in CA2 and Sncg, certainly driven by the high Nr3c2 expression.
We conclude that overall male and female mice have highly similar gene expression profiles for sex hormone receptors, and that co‐expression of sex‐ and stress hormone receptors is highly cell type specific.
3.5. Nr3c1 and Nr3c2 co‐expression with AF‐2 coregulators suggests cell type‐specific transcriptional modulation of GC signaling
Transcriptional coactivators and corepressors are key regulators of GC‐driven gene transcription. The presence of one particular coregulator can determine the outcome of GC signaling in a cell population. 35 , 36 , 37 , 38 In an in vitro screening assay, evidence was reported of 24 coregulators interacting with corticosteroid nuclear receptors: five with both receptor types, 17 with GR only and two with MR only. 24 In scRNA‐seq data, each of these coregulators showed a specific expression pattern throughout different hippocampal cell types. For example, somewhat unexpectedly, Ncoa2 was expressed in all cell types, 39 and its highest expression level was found in microglia, whereas Prox1 was mainly expressed in Vip GABAergic neurons and in the DG, where it was significantly enriched (log2FC = 1.47, p < .001) (Figure 5A; see also Table S1).
FIGURE 5.
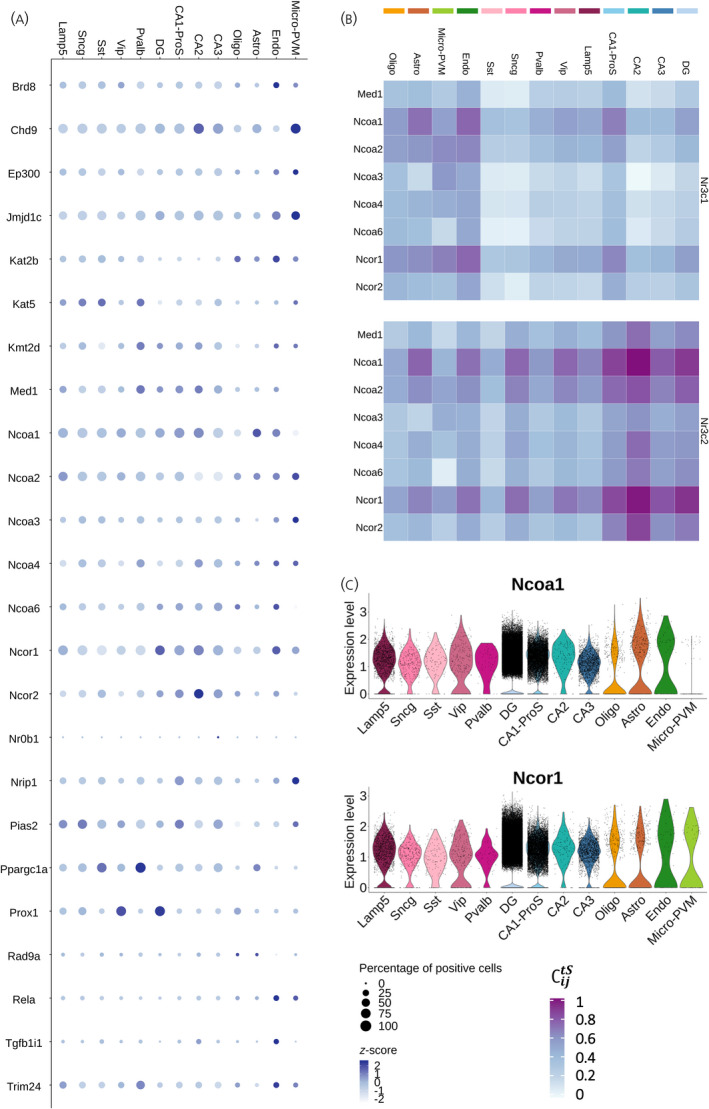
Cell type specificity of Nr3c1 and Nr3c2 co‐expression with nuclear receptor coregulators. (A) Dot plot representing both the centered log‐normalized average expression (z‐score) and the percentage of positive cells for 24 nuclear receptor AF‐2 coregulators known to interact with GR and/or MR according to an in vitro interaction screening assay. 24 (B) Heatmap representing the coupling score of Nr3c1 and Nr3c2 with a subset of GR and MR coactivators and corepressors in mouse hippocampal cell types. (C) Violin plots representing the log‐normalized expression (Expression level) of the coactivator Ncoa1 and the corepressor Ncor1 in mouse hippocampal cell types. Abbreviations: Nr3c1, nuclear receptor subfamily 3 group C member 1 (glucocorticoid receptor); Nr3c2, nuclear receptor subfamily 3 group C member 2 (mineralocorticoid receptor); Astro, astrocytes; Oligo, oligodendrocytes; Endo, endothelial cells; micro‐PVM, microglia/perivascular macrophages; Lamp5, lysosomal associated membrane protein family member 5; Vip, vasoactive intestinal peptide; Pvalb, parvalbumin; Sncg, synuclein gamma; Sst, somatostatin; DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3; AF‐2, ligand‐dependent transactivation domain 2 (helix 12); Med1, mediator complex subunit 1; Ncoa1, nuclear receptor coactivator 1; Ncoa2, nuclear receptor coactivator 2; Ncoa3, nuclear receptor coactivator 3; Ncoa4, nuclear receptor coactivator 4; Ncoa6, nuclear receptor coactivator 6; Ncor1, nuclear receptor corepressor 1; Ncor2, nuclear receptor corepressor 2
We further assessed co‐expression of AF‐2 coregulators with Nr3c1 and Nr3c2 (see Figure S2A and Table S3) for a subset of well‐characterized coactivators (Med1 and Ncoa family) and corepressors (Ncor1 and Ncor2) (Figure 5B). There was a clear co‐expression with the coregulators in non‐neuronal cells for Nr3c1 and in glutamatergic neurons for Nr3c2. Interestingly, both Nr3c1 and Nr3c2 strongly co‐expressed with Ncoa1 and Ncor1, which showed the exact same pattern of co‐expression throughout cell types. Ncoa1 and Ncor1 showed the highest coupling scores with Nr3c1 and Nr3c2 in CA1‐ProS, astrocytes and endothelial cells, and with Nr3c2 in other glutamatergic neurons, as well as Vip and Sncg GABAergic neurons (Figure 5B). Ncoa1 and Ncor1 were expressed almost at the same level in all hippocampal cell types; except for microglia, which did not express Ncoa1 (Figure 5C). Therefore, the co‐expression of these co‐regulators with stress hormone receptors is mainly driven by the cell specificity of Nr3c1 and Nr3c2 expression, with the notable exception of microglia.
3.6. Neurotransmitter and neuropeptide receptors differential co‐expression with Nr3c1 and Nr3c2 suggests synapse‐specific inputs
We next focused on neurotransmitter and neuropeptide pathways in the hippocampal glutamatergic tri‐synaptic path, which is the best characterized synaptic transmission route in the hippocampus. In this glutamatergic circuit, excitatory projections from the entorhinal cortex reach the DG granule cells through the perforant path, and the DG mossy fibers project to CA3 pyramidal neurons, which in turn stimulate CA1 neurons through the Schaffer collateral pathway. 40 In addition to the tri‐synaptic path, CA1 also receive direct and strong excitatory projections from CA2. 41 Although the sensory information mostly arrives in the DG, the CA‐regions also receive inputs from other brain regions. Afferent synapses to the tri‐synaptic path are not only glutamatergic, but also include neurotransmitters such as noradrenaline (NA), dopamine (DA) or serotonin (5‐hydroxytryptamine, 5‐HT), acetylcholine (ACh) and neuropeptides. We addressed the co‐expression of genes coding for NA, DA, 5‐HT, ACh and 33 neuropeptide receptors with Nr3c1 and Nr3c2 (Table S3), to determine how these pathways could interact with GC signaling in the hippocampal tri‐synaptic circuit.
NA receptors were mainly of the alpha‐1a, alpha‐2a/c and beta‐1 types. They co‐expressed with Nr3c1 in CA1‐ProS, and also with Nr3c2 in CA2, CA3 and the DG (Figure 6A, NA). For DA receptors, Drd5 co‐expressed strongly with Nr3c1 in CA1‐ProS and with Nr3c2 co‐expressed in all glutamatergic neurons. Drd1 co‐expressed with Nr3c2 in CA2 and the DG (Figure 6A, DA). Many 5‐HT receptors were strongly co‐expressed with Nr3c1 or Nr3c2 in all regions of the tri‐synaptic circuit, particularly Htr1a, Htr2a, Htr2c and Htr4 (Figure 6A, 5‐HT). The most consistent co‐expressed ACh receptors throughout the tri‐synaptic circuit were Chrm1 and Chrm3 (Figure 6A, Ach). Neuropeptide Y (NPY) receptors 1, 2 and 5 were strongly co‐expressed with Nr3c2 in all cell types, whereas they were more specific to CA1‐ProS and the DG for Nr3c1, which reflects specificity of steroid receptors more than of these three types of NPY receptors. Sstr2 and Sstr3 were the most co‐expressed somatostatin receptors, whereas Vipr1 was the most strongly co‐expressed vasoactive intestinal peptide receptor. Adcyap1r1 (pituitary adenylate cyclase‐activating polypeptide type I receptor) was highly co‐expressed with Nr3c1 in CA1‐ProS and with Nr3c2 in all glutamatergic neurons. Tachykinin receptor Tacr3, opioid receptor Oprl1 and corticotropin‐releasing hormone (CRH) receptor Crhr2 were co‐expressed the strongest with Nr3c1 in CA1‐ProS. Nr3c2 co‐expressed with tachykinin, arginine‐vasopressin, oxytocin, opioid, thyrotropin‐releasing hormone, relaxin, neurotensin and CRH receptors in several glutamatergic synapses (Figure 6A, Neuropeptides).
FIGURE 6.
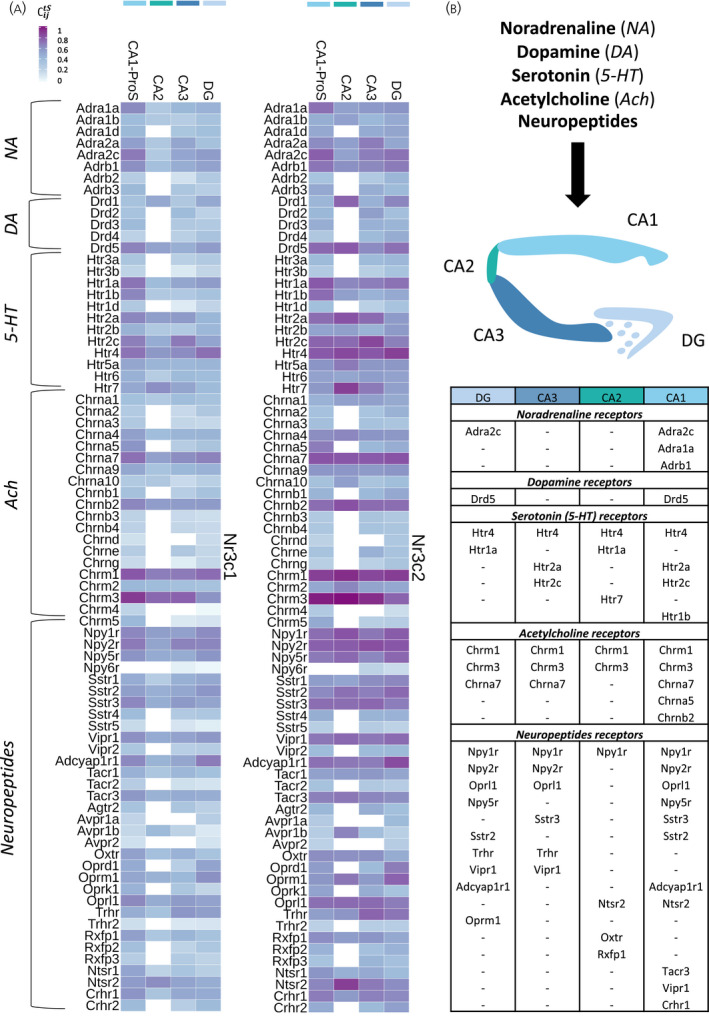
Cell type specificity of Nr3c1 and Nr3c2 co‐expression with neurotransmitter and neuropeptide receptors in the hippocampal tri‐synaptic pathway. (A) Heatmap representing the coupling score of Nr3c1 and Nr3c2 with adrenergic; dopaminergic; serotoninergic; cholinergic and neuropeptides receptors in excitatory neurons of the hippocampal tri‐synaptic pathway. (B) Table of the neurotransmitter and neuropeptide receptors above threshold in terms of coupling with Nr3c1 and Nr3c2 expression (coupling score > 0.6). Abbreviations: Nr3c1, nuclear receptor subfamily 3 group C member 1 (glucocorticoid receptor); Nr3c2, nuclear receptor subfamily 3 group C member 2 (mineralocorticoid receptor); DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3; NA, noradrenaline; DA, dopamine, 5‐HT, 5‐hydroxytryptamine; ACh, scetylcholine
We selected for the genes that had a coupling score above 0.6 both with Nr3c1 or Nr3c2 to obtain an overview of the strongest correlated neurotransmitter and neuropeptide receptors with GC signaling in the tri‐synaptic circuit (Figure 6B). For example, NA receptors are most robustly co‐expressed with Nr3c1 and Nr3c2 in the DG and CA1‐ProS.
Neurotransmitter and neuropeptide receptors co‐expression with corticosteroid receptors was more selective in GABAergic neurons and non‐neuronal cells. For example, in microglia, Nr3c1 (and Nr3c2) showed high co‐expression with Adrb1 and Adrb2. The coupling score with Ntsr2 was particularly high in astrocytes (see Figure S2B).
3.7. Nr3c1 and Nr3c2 escape de novo GRN analysis
It is known that cell‐specific gene regulation relies essentially on coordination of the activity of TFs. 42 Recent progress in high‐throughput sequencing allows the reconstruction of TF downstream networks. We applied the pySCENIC pipeline to determine whether we could identify putative MR and GR dependent regulatory networks in particular cell types. 15 The pySCENIC workflow is divided into three steps: first, it computes co‐expression modules of a TF with all correlated genes based on the scRNA‐seq count matrix. Then, these co‐expression modules are further refined by selecting genes with the TF‐specific DNA motif in their promoter region, generating the GRN modules. Finally, the refined GRN activity is measured in each individual cell, by scoring the component gene expression per GRN, and is used for new clustering (Figure 7A).
FIGURE 7.
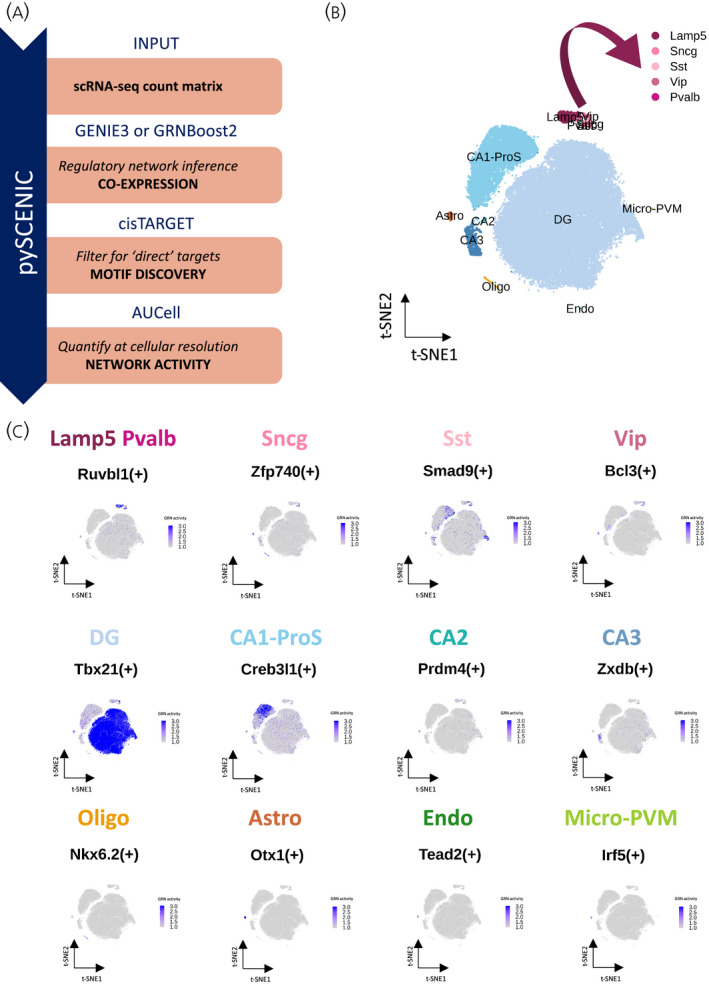
Mouse adult hippocampus gene regulatory networks (GRNs). (A) Description of the pySCENIC pipeline. (B) Dimensional reduction (t‐SNE) representation of mouse hippocampal cells grouped based on GRN activity similarities and assigned to known cell types. (C) t‐SNE representation of each hippocampal cell population most active GRN activity level per cell; scaled from 1 to 3. The sign (+) allows the distinction between a transcription factor gene (e.g., Neurod2) and this same transcription factor network; e.g Neurod2(+). Abbreviations: scRNA‐seq, single‐cell RNA sequencing; GRN, gene regulatory network; Astro, astrocytes; Oligo, oligodendrocytes; Endo, endothelial cells, micro‐PVM, microglia/perivascular macrophages; Lamp5, lysosomal associated membrane protein family member 5; Vip, vasoactive intestinal peptide; Pvalb, parvalbumin; Sncg, synuclein gamma; Sst, somatostatin; DG, dentate gyrus; CA1‐ProS, cornus ammonis 1‐prosubiculum; CA2, cornus ammonis 2; CA3, cornus ammonis 3
In this analysis, we based the t‐SNE dimensional reduction on GRN activity, rather than gene expression. The t‐SNE included the same 13 cell types, but the clustering grouped the cells differently. The most notable change was the disappearance of GABAergic neurons specificities. These neurons grouped together as one cluster, which means that all GABAergic neuronal types have very similar GRN activity profile (Figure 7B), as described previously using pySCENIC in scRNA‐seq brain data. 15 , 43 During the refinement of co‐expression modules into GRNs, the co‐expression modules with less than 80% of genes containing a binding site for the TF in their promoter region were excluded. Nr3c1 and Nr3c2 GRN activity could not be calculated as a result of not passing this threshold of motif discovery. Nevertheless, the GRN analysis allowed the identification of some cell type‐specific gene networks in the mouse hippocampus (see Figure S3A and Table S4). For example, the neuronal GRN Hsf3(+), the GABAergic GRN Maf(+) and the glutamatergic GRN Neurod2(+) showed cell type specific activity (Figure S3B). To further characterize the mouse hippocampus cell diversity, we performed a differential activity analysis on GRNs to identify the most active GRN for each cell type (Figure 7C; see also Table S5). GRNs were more specific in non‐neuronal cells. For example, Otx1(+) is the most active GRN in astrocytes, being expressed in 94% of astrocytes and only 1% of all‐other cells, with an activity enrichment log‐fold change of 4.24 (see Table S5).
Although we could not determine genes involved in Nr3c1(+) and Nr3c2(+) regulatory networks and their differential activity in hippocampal cell types, the pySCENIC allowed for a better characterization of other TF downstream networks in mouse hippocampus. This can in turn be important in determining the cellular context of stress hormone receptor activity.
4. DISCUSSION
We set out to describe the cell‐specific gene expression in the hippocampus aiming to better understand MR and GR‐mediated signaling. In a non‐treated context, corticosteroid receptor genes Nr3c1 (GR) and Nr3c2 (MR), classic GC responsive genes and newly categorized target genes showed a very heterogenous basal expression throughout hippocampal cell types, and likely predicted cell type‐specific responsiveness to GC signaling activation. Furthermore, the results on co‐expression suggested cell type‐specific crosstalk between sex and stress hormones, as well as a possible cell type‐specific transcriptional coregulation. Our results also summarize the heterogeneity in stress hormone receptor co‐expression with neurotransmitter and neuropeptide receptors in the tri‐synaptic hippocampal circuit. Finally, despite providing no further insight on GR and MR downstream GRN cell specificity, the pySCENIC pipeline revealed the cell‐specific activity of 376 TF GRNs in the mouse hippocampus. These later results further emphasize the hippocampal cell heterogeneity in terms of gene transcription activity.
Our results confirm high MR mRNA expression in glutamatergic neurons (Figure 2D), in line with its previously reported presence, and its role in mediating effects in hippocampal pyramidal and granule cell excitability. 44 , 45 , 46 , 47 , 48 MR expression in CA2 glutamatergic cells stands out, and a recent study showed that neuronal MR deletion resulted in the disappearance of CA2 molecular identity. 49 It is interesting to note that GABAergic neurons have appreciable levels of MR mRNA. To date, based on predominant presence in the granular and pyramidal cell layers, the glutamatergic cells have received most attention. However, the widespread presence of MR challenges the notion of purely cell‐autonomous effects in glutamatergic neurons. This expands the focus of future work looking into the basis of the MR‐mediated effects on cognitive and emotional processing. 50 , 51 On the other hand, MR binding to DNA earlier was linked to NeuroD factors, and this appears to reflect mainly glutamatergic neurons (see Figure S1B,C). Immunohistochemical co‐expression studies will therefore be a valuable addition to this, as well as other findings at the mRNA level.
Our data for GR also validate some known notions, such as the relatively low expression of GR mRNA in CA3 pyramidal cells (Figure 2D). 52 , 53 The presence of both receptor types in the glutamatergic CA1 neurons fits well with GR and MR cell‐autonomous opposite effects in CA1. 54 GR is certainly expressed in DG granule cells, although the percentage of positive cells is, perhaps surprisingly, modest. This may explain why corticosterone‐sensitivity of DG excitability and gene expression is markedly different from CA1 pyramidal neurons. 55 , 56 , 57 The DG is arguably the most complex structure in the hippocampus in terms of cellular diversity and organization. 58 A possible reason for the DG heterogeneity is hippocampal neurogenesis, leading to cells in different stages of neuronal maturation. Recent results suggest that neuronal progenitor cells and their progeny have intrinsic GC sensitivity and display a dorsoventral differential response to long‐term GC exposure. 59 These results could explain the contrast that we observed in MR expression. The data supported differential GC sensitivity in the DG but did not allow further subdivision in DG cells because of their overall very similar pattern of gene expression. The level of clustering that we used in the deeper analysis of the DG divided the region in only six subclusters. It is likely that more depth in the scRNA‐seq associated with clustering based on neurogenesis markers would provide further insights on MR expression in neurons at different maturation stages.
GR mRNA expression was also high in oligodendrocytes, astrocytes, microglia and endothelial cells (Figure 2D). Functionality of GR in glial cell types has previously been established, for example with cell type‐specific knockout mouse models. 60 , 61 , 62 Indeed, in a mouse model for Cushing’s disease (AdKO), we observed clear changes for astrocytes, microglia and oligodendrocytes. 63 For all of these cell types, effects of GCs, stress and/or GR antagonists (direct and indirect) have been reported in rodents and human studies. 64 , 65 , 66 , 67 Specifically, microglial cells are clearly responsive to stress and GCs, and have recently been reported to play a role in synaptic plasticity. 68 , 69 Interestingly, the signaling repertoire of GR in microglia is unique for the brain, in that Ncoa1 (coding for the steroid receptor coactivator‐1 or SRC‐1) is hardly expressed, and Ncoa2 (coding for the SRC‐2/GRIP1) may be a predominant GR coregulator (Figure 5A), analogous to immune‐modulatory GR effects in the periphery. 70 , 71 A cell type‐specific coregulator repertoire may allow more selective targeting of GR using selective receptor modulators that distinguish between downstream signaling pathways. 35 , 36 , 37 , 38 For example, in an epilepsy model, treatment with the selective GR modulator CORT108297 limited reactive microgliosis in the mouse DG without affecting an increase in astrogliosis. 72
The set of MR/GR target genes used in the present study relied on previous studies that all addressed brain or neuronal tissue. Yet, there were many differences in species, genetic background and age, exact tissue, type of intervention, dosage and type of GC used, and latency between treatment and sample collection (see Table S2). We could not provide a complete description of the conditions across the studies because they sometimes failed to mention housing and light cycle conditions, the animal sex or the timing of their intervention. Therefore, although we trust our criteria selected robustly responding GC target genes, the list is by no means exhaustive. Expression of MR/GR target genes clearly differed between cell types, but basal expression does not necessarily reflect the cell type‐specific GC responsiveness. For example, Sgk1 is known to be strongly and apparently quite selectively induced in white matter. 73 , 74 However, our results showed that Sgk1 basal mRNA levels are high in all neuronal cell types, oligodendrocytes and microglia (Figure 3A). This is an example of a gene where basal expression does not fully correlate with MR and/or GR mediated effects. However, only very few target genes show such almost binary on–off responses after GC elevations. Therefore, we expect that increased levels of Fkbp5 mRNA reflect responses in glutamatergic neurons, and those of Tsc22d3 mRNA mainly responses in other cell types. An additional argument in favor of basal expression predicting “target‐ness” is that an increased mRNA level in a relatively small cell population will be diluted by steadily high expression levels in other more abundant cell types. However, this all remains to be confirmed based on experimental data addressing responses in specific cell types. The uncertainty of cell‐specific target genes applies to a lesser extent for genes that are downregulated because this can only occur in cell types that initially expressed the gene of interest. Specific expression of downregulated genes appears to concern mainly non‐neuronal cell types (Figure 3C; see Figure S1D), for microglia clearly pointing to GR rather than MR‐mediated responses.
Susceptibility and prevalence of stress‐related neuropsychiatric and neurodegenerative pathologies differ between men and women 75 , and the prevalence of these stress‐related disorders increases in females upon drastic hormonal changes. 76 Many of these disorders have been associated with altered structure, function and neurogenic processes within the hippocampus, 77 , 78 , 79 , 80 , 81 suggesting a possible sex dimorphism in GC effects on hippocampal function. Our results showed that cell‐specific GR and MR mRNA levels correlated substantially with AR and PR mRNA (Figure 4C). This could suggest a direct crosstalk between those receptors because AR and PR can bind to GREs. 82 On the other hand, interactions with ER likely do not have a great impact in the hippocampus, given the low expression of Esr1 and Esr2 (Figure 4A and C). Thus, the quite large literature on estrogen effects on hippocampal function 83 , 84 , 85 points to involvement of membrane estrogen receptors 86 , 87 and/or interactions in afferent brain areas.
The hippocampal tri‐synaptic path receives various inputs from other brain regions and harbors a large diversity of synapses with receptors for NA, DA, 5‐HT, ACh and neuropeptides. In our results, CA1 showed the highest number of NA, DA, 5‐HT and ACh receptors that were strongly co‐expressed with GR and MR (Figure 6B). Previous studies showed that NA, DA and 5‐HT can suppress the perforant path input to CA1 by reducing postsynaptic potentials. 88 This suggests a possible interaction between GR/MR and neurotransmitter receptor signaling that could influence CA1 synaptic activity, conforming with the early work by Joëls et al. 100 Basal forebrain cholinergic neurons that project to the hippocampus are involved in stress adaptation and cognition. 89 The cholinergic system interacts with GC signaling in processes such as hippocampal‐dependent memory reconsolidation. 90 Our results suggest that the ACh receptors likely to be involved in this crosstalk are Chrm1, Chrm3 and Chrna7 (Figure 6B). In humans, higher NPY levels in serum and plasma were correlated with adaptive coping following stress as well as PTSD resilience. 92 , 93 , 94 A study in rats suggested that NPY interneuron activation in the DG contributed to trauma resilience in a model for PTSD. 94 Our results suggest that Npy1r, Npy2r and Npy5r expression is highly coupled with GR and MR mRNA levels in the DG (Figure 6B). Conceivably, NPY and GC signaling communicate via interaction of those receptors in the rodent DG (inter)neurons. Hippocampal oxytocin was found to be important for social discrimination, 95 and oxytocin can prevent stress‐induced hippocampal synaptic dysfunction and impairment of long‐term potentiation and memory. 96 Our results suggest that oxytocin signaling interference with GC signaling is mainly restrained to the hippocampal cornu ammonis region (Figure 6B). Our data also confirm the predominant role of CA2 specific AVPR1B receptors in stress‐related signaling, in conjunction with MR (Figure 6A). 49 , 98
Glucocorticoid receptor and MR activation may affect neuronal development, 99 as exemplified in CA2 pyramidal cells for MR 49 and the DG granule cells for GR. 100 This may be linked to corresponding downstream regulatory pathways. However, when looking for transcriptional networks, GR and MR did not meet the selective criteria for the pipeline motif discovery because their co‐expression modules had <80% of genes with a detected binding site in their promoter region. The pySCENIC motif discovery is limited to 10 kb down‐ and upstream of gene transcription start sites, whereas GR (and supposedly MR) binding sites are often further from their target gene start sites. 21 For hippocampal target genes, an in silico GRE interspecies screening of GC‐responsive genes showed that GREs were between 30 kb downstream and 175 kb upstream of transcription start sites of GR target gene start site, with a typical example being Adra1b that is co‐expressed with GR in pyramidal cells (Figure 6A). 23 In addition, the inability for the pySCENIC pipeline to detect MR network may have been related to an overestimation of potential MR target genes. MR mRNA levels were high in most cells in the hippocampus and significantly correlated with a total of 7319 genes. Consequently, its direct genomic targets may have been diluted by other correlated genes, leading to loss of statistical power. Nevertheless, the dominant co‐expression modules provided the cellular context in which MR and GR can bind to chromatin, and this may well be relevant, as exemplified by the Neurod2(+) GRN that may be linked to MR target genes (see Figure S3B).
Although our data in part recapitulate previous published transcriptomic studies, the cell type‐specific expression of genes that potentially interact with MR and GR allows for a reinterpretation of GC signaling in the adult mouse hippocampus. With the lack of an actual single cell transcriptomic dataset after GC treatment, the cell type‐specific expression of MR/GR downstream targets suggests gene markers to study the responsiveness of particular cell types. Moreover, the co‐expression of potentially interacting factors, such as other steroid receptors and transcriptional coregulators, defines where direct interactions can take place, and may help to more specifically target the receptors with selective modulators. 38 We hope that the results will allow the formulation of more defined future hypotheses on stress hormone effects on hippocampal function.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
Eva M. G. Viho: Conceptualization; data curation; formal analysis; investigation; methodology; project administration; visualization; writing – original draft. Jacobus C. Buurstede: Data curation; formal analysis; writing – review and editing. Jari B. Berkhout: Data curation; formal analysis; writing – review and editing. Ahmed Mahfouz: Conceptualization; investigation; methodology; supervision; writing – review and editing. Onno C. Meijer: Conceptualization; funding acquisition; investigation; project administration; supervision; writing – review and editing.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/jne.13072.
OPEN RESEARCH BADGES
This article has been awarded Open Materials, Open Data Badges. All materials and data are publicly accessible via the Open Science Framework at https://github.com/eviho/10XHip2021_VihoEMG.git.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1
Table S2
Table S3
Table S4
Table S5
ACKNOWLEDGMENTS
We thank Eduardo Umeoka, Marcia Umeoka and Harm Krugers for their involvement in the animal experiment followed by RNA sequencing of the mouse ventral hippocampus, as well as Thies Gehrmann and Daniele Bizzarri for their help with the bioinformatics. Eva M. G. Viho was employed via Corcept Therapeutics funding to Onno C. Meijer. Ahmed Mahfouz’ contribution was partially supported by an NWO Gravitation project: BRAINSCAPES: A Roadmap from Neurogenetics to Neurobiology (NWO: 024.004.012).
Viho EMG, Buurstede JC, Berkhout JB, Mahfouz A, Meijer OC. Cell type specificity of glucocorticoid signaling in the adult mouse hippocampus. J Neuroendocrinol. 2022;34:e13072. doi: 10.1111/jne.13072
DATA AVAILABILITY STATEMENT
The bulk RNA‐seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE184924. The code that supports the findings of this study is openly available in the GitHub repository (https://github.com/eviho/10XHip2021_VihoEMG). The datasets used in the code can be downloaded from Zenodo (https://doi.org/10.5281/zenodo.5729701).
REFERENCES
- 1. de Kloet ER, Joëls M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6(6):463‐475. doi: 10.1038/nrn1683 [DOI] [PubMed] [Google Scholar]
- 2. Joëls M. Corticosteroids and the brain. J Endocrinol. 2018;238(3):R121‐R130. doi: 10.1530/JOE-18-0226 [DOI] [PubMed] [Google Scholar]
- 3. Goldfarb EV, Rosenberg MD, Seo D, Constable RT, Sinha R. Hippocampal seed connectome‐based modeling predicts the feeling of stress. Nat Commun. 2020;11(1):2650. doi: 10.1038/s41467-020-16492-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kalafatakis K, Russell GM, Lightman SL. Mechanisms in endocrinology: does circadian and ultradian glucocorticoid exposure affect the brain? Eur J Endocrinol. 2019;180(2):R73‐R89. doi: 10.1530/EJE-18-0853 [DOI] [PubMed] [Google Scholar]
- 5. Fietta P, Fietta P, Delsante G. Central nervous system effects of natural and synthetic glucocorticoids. Psychiatry Clin Neurosci. 2009;63(5):613‐622. doi: 10.1111/j.1440-1819.2009.02005.x [DOI] [PubMed] [Google Scholar]
- 6. Quaedflieg CWEM, Schwabe L. Memory dynamics under stress. Mem Hove Engl. 2018;26(3):364‐376. doi: 10.1080/09658211.2017.1338299 [DOI] [PubMed] [Google Scholar]
- 7. Reul JMHM, Kloet ERD. Two receptor systems for corticosterone in rat brain: microdistribution and differential occupation. Endocrinology. 1985;117(6):2505‐2511. doi: 10.1210/endo-117-6-2505 [DOI] [PubMed] [Google Scholar]
- 8. Zeisel A, Hochgerner H, Lönnerberg P, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174(4):999‐1014.e22. doi: 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saunders A, Macosko EZ, Wysoker A, et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174(4):1015‐1030.e16. doi: 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hodge RD, Bakken TE, Miller JA, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573(7772):61‐68. doi: 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao Z, van Velthoven CTJ, Nguyen TN, et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell. 2021;184(12):3222‐3241.e26. doi: 10.1016/j.cell.2021.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mahfouz A, Lelieveldt BPF, Grefhorst A, et al. Genome‐wide coexpression of steroid receptors in the mouse brain: identifying signaling pathways and functionally coordinated regions. Proc Natl Acad Sci USA. 2016;113(10):2738‐2743. doi: 10.1073/pnas.1520376113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fiers MWEJ, Minnoye L, Aibar S, Bravo González‐Blas C, Kalender Atak Z, Aerts S. Mapping gene regulatory networks from single‐cell omics data. Brief Funct Genomics. 2018;17(4):246‐254. doi: 10.1093/bfgp/elx046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aibar S, González‐Blas CB, Moerman T, et al. SCENIC: single‐cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083‐1086. doi: 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van de Sande B, Flerin C, Davie K, et al. A scalable SCENIC workflow for single‐cell gene regulatory network analysis. Nat Protoc. 2020;15(7):2247‐2276. doi: 10.1038/s41596-020-0336-2 [DOI] [PubMed] [Google Scholar]
- 16. Tasic B, Yao Z, Graybuck LT, et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563(7729):72‐78. doi: 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single‐cell data. Cell. 2019;177(7):1888‐1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glucocorticoids JGR. Glucocorticoids, genes and brain function. Prog Neuropsychopharmacol Biol Psychiatry. 2018;82:136–168. [DOI] [PubMed] [Google Scholar]
- 20. van Weert LTCM, Buurstede JC, Mahfouz A, et al. NeuroD factors discriminate mineralocorticoid from glucocorticoid receptor DNA binding in the male rat brain. Endocrinology. 2017;158(5):1511‐1522. doi: 10.1210/en.2016-1422 [DOI] [PubMed] [Google Scholar]
- 21. Buurstede JC, van Weert LTCM, Colucci P, et al. Hippocampal glucocorticoid target genes associated with enhancement of memory consolidation. Eur J Neurosci 2021. doi: 10.1111/ejn.15226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. So AY‐L, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor‐binding sequences at glucocorticoid‐induced genes. Proc Natl Acad Sci USA. 2008;105(15):5745‐5749. doi: 10.1073/pnas.0801551105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Datson NA, Polman JAE, de Jonge RT, et al. Specific regulatory motifs predict glucocorticoid responsiveness of hippocampal gene expression. Endocrinology. 2011;152(10):3749‐3757. doi: 10.1210/en.2011-0287 [DOI] [PubMed] [Google Scholar]
- 24. Broekema MF, Hollman DAA, Koppen A, et al. Profiling of 3696 nuclear receptor‐coregulator interactions: a resource for biological and clinical discovery. Endocrinology. 2018;159(6):2397‐2407. doi: 10.1210/en.2018-00149 [DOI] [PubMed] [Google Scholar]
- 25. Smith SJ, Sümbül U, Graybuck LT, et al. Single‐cell transcriptomic evidence for dense intracortical neuropeptide networks. Ginty DD, Marder E, Fishell G, Sabatini BL, Banghart MR, eds. eLife. 2019;8:e47889. doi: 10.7554/eLife.47889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kennedy CL, Carter SD, Mifsud KR, Reul JMHM. Unexpected effects of metyrapone on corticosteroid receptor interaction with the genome and subsequent gene transcription in the hippocampus of male rats. J Neuroendocrinol. 2020;32(2):e12820. doi: 10.1111/jne.12820 [DOI] [PubMed] [Google Scholar]
- 27. Frodl T, Carballedo A, Frey E‐M, et al. Expression of glucocorticoid inducible genes is associated with reductions in cornu ammonis and dentate gyrus volumes in patients with major depressive disorder. Dev Psychopathol. 2014;26(4pt2):1209‐1217. doi: 10.1017/S0954579414000972 [DOI] [PubMed] [Google Scholar]
- 28. van Zuiden M, Geuze E, Willemen HLDM, et al. Glucocorticoid receptor pathway components predict posttraumatic stress disorder symptom development: a prospective study. Biol Psychiatry. 2012;71(4):309‐316. doi: 10.1016/j.biopsych.2011.10.026 [DOI] [PubMed] [Google Scholar]
- 29. Sarabdjitsingh RA, Isenia S, Polman A, et al. Disrupted corticosterone pulsatile patterns attenuate responsiveness to glucocorticoid signaling in rat brain. Endocrinology. 2010;151(3):1177‐1186. doi: 10.1210/en.2009-1119 [DOI] [PubMed] [Google Scholar]
- 30. Koorneef LL, Kroon J, Viho EMG, et al. The selective glucocorticoid receptor antagonist CORT125281 has tissue‐specific activity. J Endocrinol. 2020;246(1):79‐92. doi: 10.1530/JOE-19-0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Weert LTCM, Buurstede JC, Sips HCM, et al. Identification of mineralocorticoid receptor target genes in the mouse hippocampus. J Neuroendocrinol. 2019;31(8):e12735. doi: 10.1111/jne.12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen S, Wang J, Yu G, Liu W, Pearce D. Androgen and glucocorticoid receptor heterodimer formation: a possible mechanism for mutual inhibition of transcriptional activity. J Biol Chem. 1997;272(22):14087‐14092. doi: 10.1074/jbc.272.22.14087 [DOI] [PubMed] [Google Scholar]
- 33. Miranda TB, Voss TC, Sung M‐H, et al. Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 2013;73(16):5130‐5139. doi: 10.1158/0008-5472.CAN-13-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Bosscher K, Desmet SJ, Clarisse D, Estébanez‐Perpiña E, Brunsveld L. Nuclear receptor crosstalk — defining the mechanisms for therapeutic innovation. Nat Rev Endocrinol. 2020;16(7):363‐377. doi: 10.1038/s41574-020-0349-5 [DOI] [PubMed] [Google Scholar]
- 35. Zalachoras I, Houtman R, Atucha E, et al. Differential targeting of brain stress circuits with a selective glucocorticoid receptor modulator. Proc Natl Acad Sci USA. 2013;110(19):7910‐7915. doi: 10.1073/pnas.1219411110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zalachoras I, Verhoeve SL, Toonen LJ, et al. Isoform switching of steroid receptor co‐activator‐1 attenuates glucocorticoid‐induced anxiogenic amygdala CRH expression. Mol Psychiatry. 2016;21(12):1733‐1739. doi: 10.1038/mp.2016.16 [DOI] [PubMed] [Google Scholar]
- 37. Lachize S, Apostolakis EM, van der Laan S, et al. Steroid receptor coactivator‐1 is necessary for regulation of corticotropin‐releasing hormone by chronic stress and glucocorticoids. Proc Natl Acad Sci USA. 2009;106(19):8038‐8042. doi: 10.1073/pnas.0812062106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viho EMG, Buurstede JC, Mahfouz A, et al. Corticosteroid action in the brain: the potential of selective receptor modulation. Neuroendocrinology. 2019;109(3):266‐276. doi: 10.1159/000499659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meijer OC, van der Laan S, Lachize S, Steenbergen PJ, de Kloet ER. Steroid receptor coregulator diversity: what can it mean for the stressed brain? Neuroscience. 2006;138(3):891‐899. doi: 10.1016/j.neuroscience.2005.07.004 [DOI] [PubMed] [Google Scholar]
- 40. Basu J, Siegelbaum SA. The corticohippocampal circuit, synaptic plasticity, and memory. Cold Spring Harb Perspect Biol. 2015;7(11):a021733. doi: 10.1101/cshperspect.a021733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chevaleyre V, Siegelbaum SA. Strong CA2 pyramidal neuron synapses define a powerful disynaptic cortico‐hippocampal loop. Neuron. 2010;66(4):560‐572. doi: 10.1016/j.neuron.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suo S, Zhu Q, Saadatpour A, Fei L, Guo G, Yuan G‐C. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep. 2018;25(6):1436‐1445.e3. doi: 10.1016/j.celrep.2018.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeisel A, Muñoz‐Manchado AB, Codeluppi S, et al. Cell types in the mouse cortex and hippocampus revealed by single‐cell RNA‐seq. Science. 2015;347(6226):1138‐1142. doi: 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- 44. Le Menuet D, Lombès M. The neuronal mineralocorticoid receptor: from cell survival to neurogenesis. Steroids. 2014;91:11‐19. doi: 10.1016/j.steroids.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 45. Kretz O, Schmid W, Berger S, Gass P. The mineralocorticoid receptor expression in the mouse CNS is conserved during development. NeuroReport. 2001;12(6):1133‐1137. [DOI] [PubMed] [Google Scholar]
- 46. Patel PD, Lopez JF, Lyons DM, Burke S, Wallace M, Schatzberg AF. Glucocorticoid and mineralocorticoid receptor mRNA expression in squirrel monkey brain. J Psychiatr Res. 2000;34(6):383‐392. doi: 10.1016/S0022-3956(00)00035-2 [DOI] [PubMed] [Google Scholar]
- 47. Kalman BA, Spencer RL. Rapid corticosteroid‐dependent regulation of mineralocorticoid receptor protein expression in rat brain. Endocrinology. 2002;143(11):4184‐4195. doi: 10.1210/en.2002-220375 [DOI] [PubMed] [Google Scholar]
- 48. Vázquez DM, López JF, Morano MI, Kwak SP, Watson SJ, Akil H. α, β, and γ mineralocorticoid receptor messenger ribonucleic acid splice variants: differential expression and rapid regulation in the developing hippocampus. Endocrinology. 1998;139(7):3165‐3177. doi: 10.1210/endo.139.7.6095 [DOI] [PubMed] [Google Scholar]
- 49. McCann KE, Lustberg DJ, Shaughnessy EK, et al. Novel role for mineralocorticoid receptors in control of a neuronal phenotype. Mol Psychiatry. 2021;26(1):350‐364. doi: 10.1038/s41380-019-0598-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwabe L, Tegenthoff M, Höffken O, Wolf OT. Mineralocorticoid receptor blockade prevents stress‐induced modulation of multiple memory systems in the human brain. Biol Psychiatry. 2013;74(11):801‐808. doi: 10.1016/j.biopsych.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 51. ter Heegde F, De Rijk RH, Vinkers CH. The brain mineralocorticoid receptor and stress resilience. Psychoneuroendocrinology. 2015;52:92‐110. doi: 10.1016/j.psyneuen.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 52. Morimoto M, Morita N, Ozawa H, Yokoyama K, Kawata M. Distribution of glucocorticoid receptor immunoreactivity and mRNA in the rat brain: an immunohistochemical and in situ hybridization study. Neurosci Res. 1996;26(3):235‐269. doi: 10.1016/S0168-0102(96)01105-4 [DOI] [PubMed] [Google Scholar]
- 53. Wang Q, Van Heerikhuize J, Aronica E, et al. Glucocorticoid receptor protein expression in human hippocampus; stability with age. Neurobiol Aging. 2013;34(6):1662‐1673. doi: 10.1016/j.neurobiolaging.2012.11.019 [DOI] [PubMed] [Google Scholar]
- 54. de Kloet ER, Meijer OC, de Nicola AF, de Rijk RH, Joëls M. Importance of the brain corticosteroid receptor balance in metaplasticity, cognitive performance and neuro‐inflammation. Front Neuroendocrinol. 2018;49:124‐145. doi: 10.1016/j.yfrne.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 55. van Gemert NG, Carvalho DMM, Karst H, et al. Dissociation between rat hippocampal CA1 and dentate gyrus cells in their response to corticosterone: effects on calcium channel protein and current. Endocrinology. 2009;150(10):4615‐4624. doi: 10.1210/en.2009-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pasricha N, Joëls M, Karst H. Rapid effects of corticosterone in the mouse dentate gyrus via a nongenomic pathway. J Neuroendocrinol. 2011;23(2):143‐147. doi: 10.1111/j.1365-2826.2010.02091.x [DOI] [PubMed] [Google Scholar]
- 57. Meijer OC, de Kloet ER. A role for the mineralocorticoid receptor in a rapid and transient suppression of hippocampal 5‐HT1A receptor mRNA by corticosterone. J Neuroendocrinol. 1995;7(8):653‐657. doi: 10.1111/j.1365-2826.1995.tb00804.x [DOI] [PubMed] [Google Scholar]
- 58. Amaral DG, Scharfman HE, Lavenex P. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies). In: Scharfman HE ed. Progress in brain research. Vol 163. The dentate gyrus: a comprehensive guide to structure, function, and clinical implications. Elsevier; 2007:3‐790. doi: 10.1016/S0079-6123(07)63001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Levone BR, Codagnone MG, Moloney GM, Nolan YM, Cryan JF, O’ Leary OF. Adult‐born neurons from the dorsal, intermediate, and ventral regions of the longitudinal axis of the hippocampus exhibit differential sensitivity to glucocorticoids. Mol Psychiatry. 2021;26(7):3240‐3252. 10.1038/s41380-020-0848-8 [DOI] [PubMed] [Google Scholar]
- 60. Madalena KM, Lerch JK. The effect of glucocorticoid and glucocorticoid receptor interactions on brain, spinal cord, and glial cell plasticity. Neural Plast. 2017;2017:e8640970. doi: 10.1155/2017/8640970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ros‐Bernal F, Hunot S, Herrero MT, et al. Microglial glucocorticoid receptors play a pivotal role in regulating dopaminergic neurodegeneration in parkinsonism. Proc Natl Acad Sci USA. 2011;108(16):6632‐6637. doi: 10.1073/pnas.1017820108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maatouk L, Yi C, Carrillo‐de Sauvage M‐A, et al. Glucocorticoid receptor in astrocytes regulates midbrain dopamine neurodegeneration through connexin hemichannel activity. Cell Death Differ. 2019;26(3):580‐596. doi: 10.1038/s41418-018-0150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Amaya JM, Suidgeest E, Sahut‐Barnola I, et al. Effects of long‐term endogenous corticosteroid exposure on brain volume and glial cells in the AdKO mouse. Front Neurosci. 2021;15:604103. doi: 10.3389/fnins.2021.604103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meyer JS. Early adrenalectomy stimulates subsequent growth and development of the rat brain. Exp Neurol. 1983;82(2):432‐446. doi: 10.1016/0014-4886(83)90415-6 [DOI] [PubMed] [Google Scholar]
- 65. Alonso G. Prolonged corticosterone treatment of adult rats inhibits the proliferation of oligodendrocyte progenitors present throughout white and gray matter regions of the brain. Glia. 2000;31(3):219‐231. doi: [DOI] [PubMed] [Google Scholar]
- 66. Zivadinov R, Rudick RA, Masi RD, et al. Effects of IV methylprednisolone on brain atrophy in relapsing‐remitting MS. Neurology. 2001;57(7):1239‐1247. doi: 10.1212/WNL.57.7.1239 [DOI] [PubMed] [Google Scholar]
- 67. Lou Y‐X, Li J, Wang Z‐Z, Xia C‐Y, Chen N‐H. Glucocorticoid receptor activation induces decrease of hippocampal astrocyte number in rats. Psychopharmacology. 2018;235(9):2529‐2540. doi: 10.1007/s00213-018-4936-2 [DOI] [PubMed] [Google Scholar]
- 68. Wohleb ES, Hanke ML, Corona AW, et al. β‐adrenergic receptor antagonism prevents anxiety‐like behavior and microglial reactivity induced by repeated social defeat. J Neurosci. 2011;31(17):6277‐6288. doi: 10.1523/JNEUROSCI.0450-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sanguino‐Gómez J, Buurstede JC, Abiega O, et al. An emerging role for microglia in stress‐effects on memory. Eur J Neurosci. 2021. Mar 16. doi: 10.1111/ejn.15188. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA. 2002;99(26):16701‐16706. doi: 10.1073/pnas.262671599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor‐mediated immunosuppression. EMBO J. 2006;25(1):108‐117. doi: 10.1038/sj.emboj.7600919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wulsin AC, Kraus KL, Gaitonde KD, et al. The glucocorticoid receptor specific modulator CORT108297 reduces brain pathology following status epilepticus. Exp Neurol. 2021;341:113703. doi: 10.1016/j.expneurol.2021.113703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hinds LR, Chun LE, Woodruff ER, Christensen JA, Hartsock MJ, Spencer RL. Dynamic glucocorticoid‐dependent regulation of Sgk1 expression in oligodendrocytes of adult male rat brain by acute stress and time of day. PLoS One. 2017;12(4):e0175075. doi: 10.1371/journal.pone.0175075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van Gemert NG, Meijer OC, Morsink MC, Joëls M. Effect of brief corticosterone administration on SGK1 and RGS4 mRNA expression in rat hippocampus. Stress. 2006;9(3):165‐170. doi: 10.1080/10253890600966169 [DOI] [PubMed] [Google Scholar]
- 75. Hillerer KM, Slattery DA, Pletzer B. Neurobiological mechanisms underlying sex‐related differences in stress‐related disorders: effects of neuroactive steroids on the hippocampus. Front Neuroendocrinol. 2019;55:100796. doi: 10.1016/j.yfrne.2019.100796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Slavich GM, Sacher J. Stress, sex hormones, inflammation, and major depressive disorder: Extending Social Signal Transduction Theory of Depression to account for sex differences in mood disorders. Psychopharmacology. 2019;236(10):3063‐3079. doi: 10.1007/s00213-019-05326-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Woolley CS, Gould E, Frankfurt M, McEwen BS. Naturally occurring fluctuation in dendritic spine density on adult hippocampal pyramidal neurons. J Neurosci. 1990;10(12):4035‐4039. doi: 10.1523/JNEUROSCI.10-12-04035.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Felmingham K, Williams LM, Kemp AH, et al. Neural responses to masked fear faces: sex differences and trauma exposure in posttraumatic stress disorder. ‐ PsycNET. J Abnorm Psychol. 2010;119(1):241‐247. doi: 10.1037/a0017551 [DOI] [PubMed] [Google Scholar]
- 79. Milne AMB, MacQueen GM, Hall GBC. Abnormal hippocampal activation in patients with extensive history of major depression: an fMRI study. J Psychiatry Neurosci. 2012;37(1): doi: 10.1503/jpn.110004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Holsen LM, Spaeth SB, Lee J‐H, et al. Stress response circuitry hypoactivation related to hormonal dysfunction in women with major depression. J Affect Disord. 2011;131(1):379‐387. doi: 10.1016/j.jad.2010.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Holsen LM, Lee J‐H, Spaeth SB, et al. Brain hypoactivation, autonomic nervous system dysregulation, and gonadal hormones in depression: a preliminary study. Neurosci Lett. 2012;514(1):57‐61. doi: 10.1016/j.neulet.2012.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nelson CC, Hendy SC, Shukin RJ, et al. Determinants of DNA sequence specificity of the androgen, progesterone, and glucocorticoid receptors: evidence for differential steroid receptor response elements. Mol Endocrinol. 1999;13(12):2090‐2107. doi: 10.1210/mend.13.12.0396 [DOI] [PubMed] [Google Scholar]
- 83. Shughrue PJ, Merchenthaler I. Estrogen is more than just a “sex hormone”: novel sites for estrogen action in the hippocampus and cerebral cortex. Front Neuroendocrinol. 2000;21(1):95‐101. doi: 10.1006/frne.1999.0190 [DOI] [PubMed] [Google Scholar]
- 84. Mukai H, Kimoto T, Hojo Y, et al. Modulation of synaptic plasticity by brain estrogen in the hippocampus. Biochim Biophys Acta BBA ‐ Gen Subj. 2010;1800(10):1030‐1044. doi: 10.1016/j.bbagen.2009.11.002 [DOI] [PubMed] [Google Scholar]
- 85. Wang W, Le AA, Hou B, et al. Memory‐related synaptic plasticity is sexually dimorphic in rodent hippocampus. J Neurosci. 2018;38(37):7935‐7951. doi: 10.1523/JNEUROSCI.0801-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kuroki Y, Fukushima K, Kanda Y, Mizuno K, Watanabe Y. Putative membrane‐bound estrogen receptors possibly stimulate mitogen‐activated protein kinase in the rat hippocampus. Eur J Pharmacol. 2000;400(2):205‐209. doi: 10.1016/S0014-2999(00)00425-8 [DOI] [PubMed] [Google Scholar]
- 87. Wu T‐W, Chen S, Brinton RD. Membrane estrogen receptors mediate calcium signaling and MAP kinase activation in individual hippocampal neurons. Brain Res. 2011;1379:34‐43. doi: 10.1016/j.brainres.2011.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Otmakhova NA, Lewey J, Asrican B, Lisman JE. Inhibition of perforant path input to the CA1 region by serotonin and noradrenaline. J Neurophysiol. 2005;94(2):1413‐1422. doi: 10.1152/jn.00217.2005 [DOI] [PubMed] [Google Scholar]
- 89. Joels M. Corticosteroid effects on electrical properties of brain cells: Temporal aspects and role of antiglucocorticoids. Psychoneuroendocrinology. 1997;22:S81–S86. 10.1016/s0306-4530(97)00013-9 [DOI] [PubMed] [Google Scholar]
- 90. Paul S, Jeon WK, Bizon JL, Han J‐S. Interaction of basal forebrain cholinergic neurons with the glucocorticoid system in stress regulation and cognitive impairment. Front Aging Neurosci. 2015;7:43. doi: 10.3389/fnagi.2015.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Amiri S, Jafarian Z, Vafaei AA, Motaghed‐Larijani Z, Samaei SA, Rashidy‐Pour A. Glucocorticoids interact with cholinergic system in impairing memory reconsolidation of an inhibitory avoidance task in mice. Basic Clin Neurosci. 2015;6(3):155‐162. [PMC free article] [PubMed] [Google Scholar]
- 92. Reijnen A, Geuze E, Eekhout I, et al. Biological profiling of plasma neuropeptide Y in relation to posttraumatic stress symptoms in two combat cohorts. Biol Psychol. 2018;134:72‐79. doi: 10.1016/j.biopsycho.2018.02.008 [DOI] [PubMed] [Google Scholar]
- 93. Yehuda R, Brand S, Yang R‐K. Plasma neuropeptide Y concentrations in combat exposed veterans: relationship to trauma exposure, recovery from PTSD, and coping. Biol Psychiatry. 2006;59(7):660‐663. doi: 10.1016/j.biopsych.2005.08.027 [DOI] [PubMed] [Google Scholar]
- 94. Morgan CA, Wang S, Southwick SM, et al. Plasma neuropeptide‐Y concentrations in humans exposed to military survival training. Biol Psychiatry. 2000;47(10):902‐909. doi: 10.1016/S0006-3223(99)00239-5 [DOI] [PubMed] [Google Scholar]
- 95. Regev‐Tsur S, Demiray YE, Tripathi K, Stork O, Richter‐Levin G, Albrecht A. Region‐specific involvement of interneuron subpopulations in trauma‐related pathology and resilience. Neurobiol Dis. 2020;143:104974. doi: 10.1016/j.nbd.2020.104974 [DOI] [PubMed] [Google Scholar]
- 96. Raam T, McAvoy KM, Besnard A, Veenema AH, Sahay A. Hippocampal oxytocin receptors are necessary for discrimination of social stimuli. Nat Commun. 2017;8(1):2001. doi: 10.1038/s41467-017-02173-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee S‐Y, Park S‐H, Chung C, Kim JJ, Choi S‐Y, Han J‐S. Oxytocin protects hippocampal memory and plasticity from uncontrollable stress. Sci Rep. 2015;5(1):18540. doi: 10.1038/srep18540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pagani JH, Zhao M, Cui Z, et al. Role of the vasopressin 1b receptor in rodent aggressive behavior and synaptic plasticity in hippocampal area CA2. Mol Psychiatry. 2015;20(4):490‐499. doi: 10.1038/mp.2014.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cruceanu C, Dony L, Krontira AC, et al. Cell‐type‐specific impact of glucocorticoid receptor activation on the developing brain: A Cerebral Organoid Study. American Journal of Psychiatry. 2021. doi: 10.1176/appi.ajp.2021.21010095 [DOI] [PubMed] [Google Scholar]
- 100. Fitzsimons CP, van Hooijdonk LWA, Schouten M, et al. Knockdown of the glucocorticoid receptor alters functional integration of newborn neurons in the adult hippocampus and impairs fear‐motivated behavior. Mol Psychiatry. 2013;18(9):993‐1005. doi: 10.1038/mp.2012.123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1
Table S2
Table S3
Table S4
Table S5
Data Availability Statement
The bulk RNA‐seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE184924. The code that supports the findings of this study is openly available in the GitHub repository (https://github.com/eviho/10XHip2021_VihoEMG). The datasets used in the code can be downloaded from Zenodo (https://doi.org/10.5281/zenodo.5729701).


