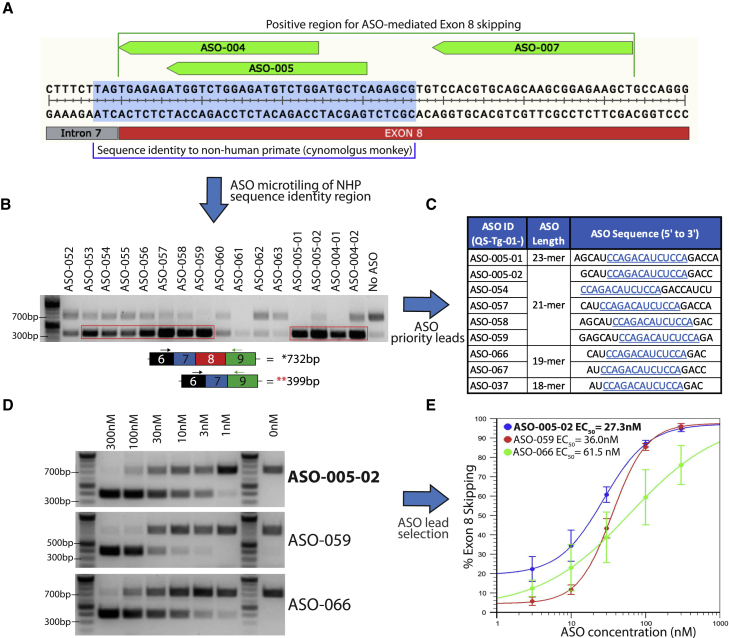
Figure 2.
ASO tiling, optimization, and selection of ASO leads that induce TECPR2 exon 8 skipping
(A) TECPR2 exon 8 region showing binding locations of the three most active exon 8-skipping ASOs (ASO-004, ASO-005, and ASO-007, shown in green). Sub-region with 100% sequence identity to cynomolgus monkey is highlighted in light blue. (B) DNA gel inverse image with a subset of PCR results of additional screening after ASO micro-tiling and trimming of initial active sequences. Enrichment of lower PCR amplicon (∼399 bp) can be detected after treatment with several of these ASOs (highlighted in red rectangle). (C) Subset of nine ASO sequences of different length selected as priority leads. All sequences listed were then synthesized using 2′-O-methoxyethyl bases and 5-methyl cytosine and uracil bases, with phosphorothioate backbone. All ASO sequences share a core 13-base sequence highlighted in blue and underscored. (D) DNA gel inverse images of TECPR2 exon 8-skipping PCR results for ASO leads ASO-005-02, ASO-059, and ASO-066, assayed at different concentrations (300–1 nM). (E) Half maximal effective concentration (EC50) for ASO leads ASO-005-02, ASO-059, and ASO-066 calculated using percentage of TECPR2 exon 8 skipping (estimated as fraction of lower PCR amplicon over total PCR product) at different concentrations (each data point corresponds to n = 3 independent rounds of ASO treatment, each with n = 1 RT-PCR assay). ASO-005-02 (in blue) was the most potent (EC50 = ∼27.3 nM) and was selected for further studies in patient-derived cells.