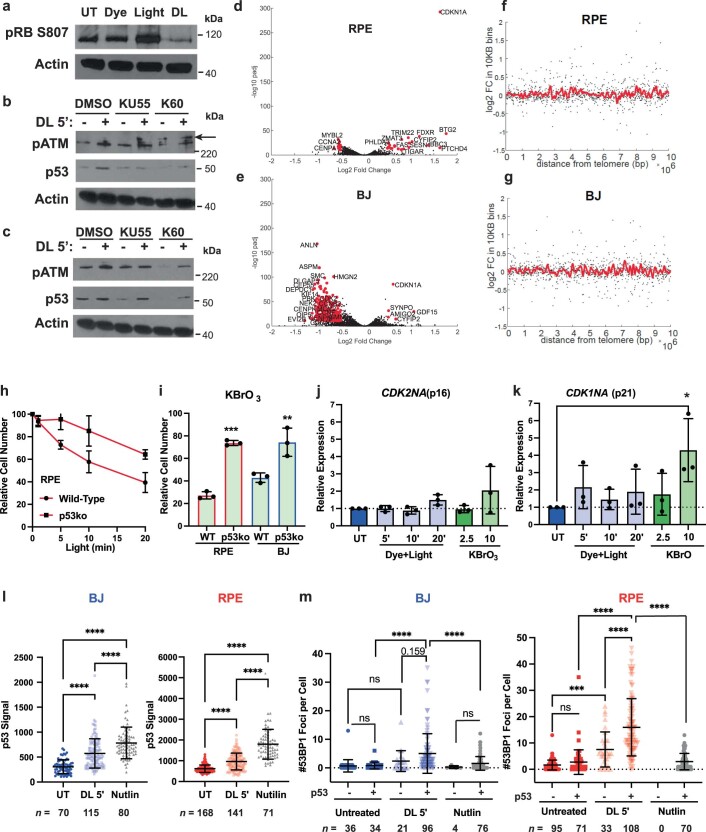
Extended Data Fig. 5. 8oxoG induced DDR signaling and p53 activation.
(a) Immunoblot of phosphorylated RB from BJ FAP-TRF1 cells 4 days after dye, light or DL for 5 min. (b, c) Immunoblots of BJ (b) and RPE (c) FAP-TRF1 cells 3 hours after 5 min DL. Cells were pre- and post-treated with ATMi KU55933 (10 μM) or KU60019 (1 μM) or DMSO. Arrow indicates non-specific band. (d, e) Volcano plots of gene expression changes in RPE (d) and BJ (e) FAP-TRF1 cells 24 hours after dye + light. Each dot is a gene and red dots are significantly up or down-regulated. HeLa cells showed no significant changes. Analyzed with DEseq2. (f, g) Gene expression analysis in RPE (f) and BJ (g) FAP-TRF1 cells 24 hours after dye + light, as a function of chromosome position. Each dot is a 10 kb bin, and the red line = the average. (h) Counts of wild-type and p53ko RPE FAP-TRF1 cells 4 days after indicated treatment. Error bars represent the mean ± s.d. from three independent experiments. (i) Counts of wild-type and p53ko RPE and BJ FAP-TRF1 cells 4 days after treatment with 10 mM (RPE) or 2.5 mM (BJ) KBrO3. (j, k) qPCR analysis of p16 mRNA (CDK2NA) and p21 mRNA (CDK1NA) in BJ FAP-TRF1 cells, 4 days after treatment. For panels i-k, error bars represent the mean ± s.d. from the number of independent experiments indicated by the black circles. Statistical analysis by one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001). (l) Quantification of p53 protein signal intensity by IF in BJ and RPE FAP-TRF1 cells 3 hours after 5 min DL or 20 μM nutlin. (m) 53BP1 foci per cell analyzed by IF from cells as treated in panel (l). p53 signal intensity by IF was used to determine p53 expression (+). For panel l and m, error bars represent the mean ± s.d. from the indicated n number of nuclei analyzed from two independent experiments. Statistical analysis by one-way ANOVA (l) or two-way ANOVA (m) (ns=not significant, ***p < 0.001, ****p < 0.0001).