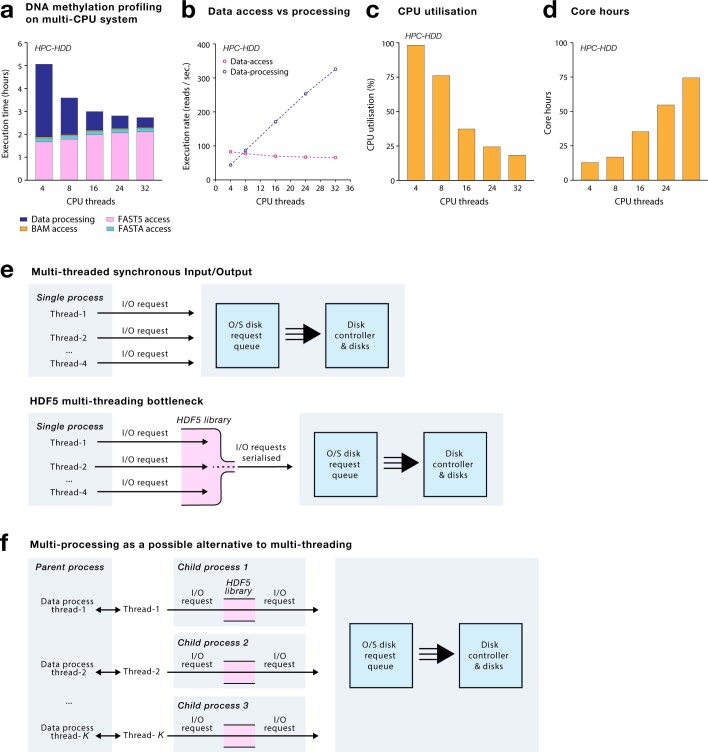
Extended Data Fig. 1. Inefficient parallel access is a major bottleneck in analysis of FAST5 files.
(a) Bar chart shows the time consumed by individual components of a Nanopolish DNA methylation calling job with signal data input in FAST5 format: FAST5 data access (pink), FASTA data access (teal), BAM data access (orange) and data processing (navy). To assess the impact of multi-threading, the analysis was run with various numbers of CPU threads on the HPC-HDD system (see Supplementary Table 2). The analysis was run on a downsampled human genome sequencing dataset of 500,000 reads (see Supplementary Table 1). (b) Dot plots show the rate of file access and processing (reads / second) during the DNA methylation calling job above, as a function of CPU threads used. (c,d) Bar charts show the proportional CPU utilisation (c) and total core hours (d) during the DNA methylation calling jobs above. The definition of core-hours is provided in the Methods section. (e) The upper schematic illustrates the architecture of a job with multi-threaded synchronous file access (I/O). The lower schematic illustrates the bottleneck created by the HDF5 library that is required to read FAST5 files. The HDF5 library serialises I/O requests, making multi-threaded analysis highly inefficient and causing the observed decline in CPU utilisation with increasing numbers of CPU threads. (f) Schematic illustrates the architecture of a multi-processing approach that was implemented to circumvent this limitation in the HDF5 library. The multi-processing approach is viable but requires challenging software engineering and is not a generalisable, long-term solution.