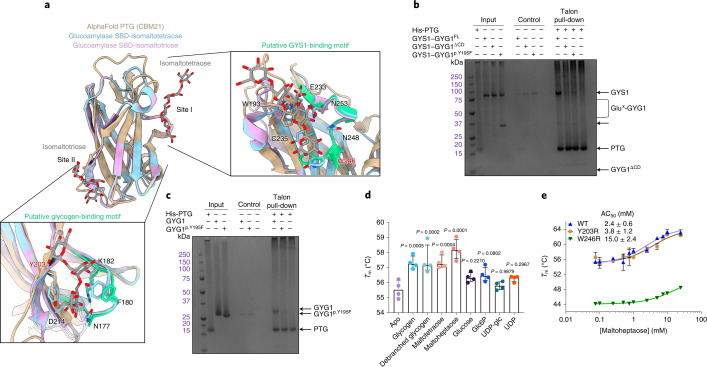
Fig. 6. CBM21 domain of PTG binds to the GYS1–GYG1 complex via the associated glucose chain.
a, Structural alignment of the AlphaFold predicted structure of the PTG(CBM21) domain against the starch-binding domain (SBD) from R. oryzae glucoamylase bound to maltotetraose and maltotriose at site I and site II, respectively. Panels show how site I and site II align with the putative GYS-binding motif and putative glycogen-binding motif. Both motifs are coloured green. Y203 and W246 labels are highlighted red. b, PTG(CBM21) was incubated with GYS1–GYG1FL, GYS1–GYG1pY195F or GYS1–GYG1ΔCD. The ability of PTG to bind GYS1–GYG1 complexes was assessed by affinity pull-down, followed by SDS–PAGE (n = 4 technical repeats). c, PTG(CBM21) was incubated with GYG1 or GYG1p.Y195F catalytic domain constructs, passed onto affinity resin and analysed by SDS–PAGE (n = 4 technical repeats). The GYG1 catalytic domain exists as a mixture of glucosylated states and runs at a higher apparent molecular weight in SDS–PAGE than GYG1p.Y195F, which is nonglucosylated. d, Thermal shift analysis of PTG(CBM21) in the presence of various sugars and ligands (n = 4 technical repeats). P values between the apo and plus sugar samples were determined by two-tailed unpaired t-test. e, Thermal shift analysis of PTG(CBM21) wild type (labelled WT), PTG(CBM21)p.Y203R variant (labelled Y203R) and PTG(CBM21)p.W246R variant (labelled W246R) in the presence of increasing concentrations of maltoheptaose. Median melting temperatures (Tm) and standard deviations are shown (n = 3 technical repeats).