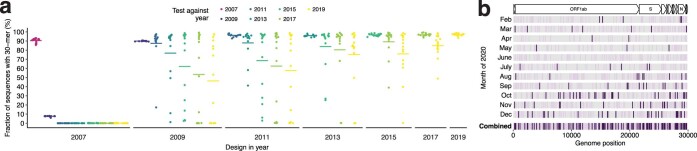
Extended Data Fig. 1. Emerging viral variation over time and the effect on diagnostic performance.
a, Diagnostic performance for influenza A virus subtyping may degrade over time, even considering conserved sites. At each year, we select the 15 most conserved 30-mers from recent sequences for segment 6 (N) for all N1 subtypes; each point is a 30-mer. Plotted value is the fraction of sequences in subsequent years (colored) that contain the 30-mer; bars are the mean. To aid visualization, only odd years are shown. 2007 N1 30-mers are absent following 2007 owing to antigenic shift during the 2009 H1N1 pandemic. b, Variation along the SARS-CoV-2 genome emerging over time during 2020. Bottom row (‘Combined’) shows all 1,131 single nucleotide polymorphisms, among 361,460 genomes, that crossed 0.1% or 1% frequency between February 1, 2020 and the end of 2020—i.e., (i) at <0.1% frequency in genomes collected before February 1 and 0.1–1% frequency by December 31 (light purple); or (ii) at <1% frequency before February 1 and at ≥1% frequency by December 31 (dark). Other rows show the month in which each polymorphism crosses the frequency threshold.