The pathogenic anti–IFN-γ autoantibodies belong to three groups based on their epitopes, leading to two signal-related neutralization modes by preventing IFN-γ binding to the receptor or IFN-γ–induced receptor dimerization. Moreover, anti–IFN-γ autoantibodies selectively induce Fc-mediated responses, potentiating their pathogenic functions.
Abstract
Anti-interferon (IFN)–γ autoantibodies (AIGAs) are a pathogenic factor in late-onset immunodeficiency with disseminated mycobacterial and other opportunistic infections. AIGAs block IFN-γ function, but their effects on IFN-γ signaling are unknown. Using a single-cell capture method, we isolated 19 IFN-γ–reactive monoclonal antibodies (mAbs) from patients with AIGAs. All displayed high-affinity (KD < 10−9 M) binding to IFN-γ, but only eight neutralized IFN-γ–STAT1 signaling and HLA-DR expression. Signal blockade and binding affinity were correlated and attributed to somatic hypermutations. Cross-competition assays identified three nonoverlapping binding sites (I–III) for AIGAs on IFN-γ. We found that site I mAb neutralized IFN-γ by blocking its binding to IFN-γR1. Site II and III mAbs bound the receptor-bound IFN-γ on the cell surface, abolishing IFN-γR1–IFN-γR2 heterodimerization and preventing downstream signaling. Site III mAbs mediated antibody-dependent cellular cytotoxicity, probably through antibody–IFN-γ complexes on cells. Pathogenic AIGAs underlie mycobacterial infections by the dual blockade of IFN-γ signaling and by eliminating IFN-γ–responsive cells.
Introduction
The implication of autoantibodies to cytokines in infectious diseases is growing, as highlighted by studies on autoantibodies against type I IFN, IL-6, and IL-17A and IL-17F underlying life-threatening coronavirus disease 2019 (COVID-19), staphylococcal disease, and mucocutaneous candidiasis, respectively (Bastard et al., 2021; Bastard et al., 2020; Ku et al., 2020; Puel et al., 2010; Puel et al., 2008). Mycobacteriosis can also be driven by anti–IFN-γ autoantibodies (AIGAs) causing a late-onset form of immunodeficiency, further expanding the spectrum of Mendelian susceptibility to mycobacterial disease (MSMD) caused by genetic inborn errors of immunity to AIGA-mediated signaling blockade (Doffinger et al., 2004; Hoflich et al., 2004; Kampmann et al., 2005; Patel et al., 2005; Shih et al., 2021). 18 genes present heterogeneous mutations in humans with MSMD: IFNG, IFNGR1, IFNGR2, STAT1, JAK1, IRF8, SPPL2A, IL12B, IL12RB1, IL12RB2, IL23R, ISG15, TYK2, RORC, CYBB, NEMO, TBX21, and ZNFX1. Most of these genes are autosomal, but two are X-linked genes (CYBB and NEMO; Boisson-Dupuis and Bustamante, 2021; Bustamante, 2020; Bustamante et al., 2014; Noma et al., 2022; Rosain et al., 2019). The study of MSMD has revealed that the production of and response to IFN-γ are crucial for antimycobacterial defenses. Like MSMD patients, individuals with AIGAs are typically susceptible to disseminated nontuberculous mycobacterial and nontyphoidal Salmonella infections, due to the ability of these antibodies to neutralize IFN-γ–induced signaling (Browne et al., 2012a; Chi et al., 2013; Chi et al., 2016). IFN-γ is a homodimeric cytokine with pleiotropic functions. It is produced principally by activated T cells, natural killer (NK) cells, and group I innate lymphoid cells in the context of stimulation with cytokines or antigen (Ivashkiv, 2018). Homodimeric IFN-γ binds to its surface receptors, which consist of two ubiquitously expressed ligand-binding chains (IFN-γR1) and two accessory chains (IFN-γR2), to form an active hexameric complex, inducing a change in conformation (Mendoza et al., 2019). However, IFN-γR2 levels may be tightly regulated to modulate the response to IFN-γ in different immune cell subsets (Bach et al., 1995). IFN-γR ligation triggers the transcription of IFN-stimulated genes (ISGs) in the nucleus via canonical JAK/STAT (Hu and Ivashkiv, 2009; Ivashkiv, 2018; Schroder et al., 2004). The IFN-γ–JAK–STAT1–ISG pathway is related to most effector functions, including chemokine production, leukocyte trafficking, antigen-presenting molecules, and antimicrobial factors (Hu and Ivashkiv, 2009; Ivashkiv, 2018; Schroder et al., 2004).
AIGAs are primarily detected in adults from Southeast Asia, with >600 patients reported since 2004 (Ku et al., 2020; Shih et al., 2021). The high prevalence of AIGAs in patients with mycobacterial infections from this region, including Taiwan and Thailand in particular, is strictly associated with two specific HLA class II haplotypes, HLA-DRB1*16:02–DQB1*05:02 and HLA-DRB1*15:02–DQB1*05:01, which confer a genetic predisposition (Chi et al., 2013; Ku et al., 2016). Indeed, most AIGA patients carry at least one allele of DRB1*16:02 or 15:02 (100% in Thailand, 93.2% in Taiwan, and 98.2% in South China; Guo et al., 2020; Ku et al., 2016). HLA class II is required to present antigens to CD4+ T cells and to prime effective immune responses and has emerged as a possible genetic source of susceptibility to autoantibodies (Reveille, 2006). The production of AIGAs therefore probably underlies the ethnic bias in disease development.
AIGAs are pathogenic because they can inhibit IFN-γ function, but their molecular mode of action remains poorly characterized. The C-terminal region of IFN-γ has been identified as a major AIGA epitope through the mapping of overlapping peptides and deletion variants of the human IFN-γ protein (Lin et al., 2016; Wipasa et al., 2018), where the C-terminal region is involved in interactions between IFN-γ and its receptor (Lundell and Narula, 1994). A C-terminally modified IFN-γ, epitope-erased IFN-γ (EE-IFN-γ), yields significantly higher levels of STAT-1 phosphorylation than WT IFN-γ in peripheral blood mononuclear cells (PBMCs) incubated with plasma containing AIGAs, confirming the role of this region in AIGA neutralization against IFN-γ (Lin et al., 2016). The major epitope of IFN-γ is a region displaying a high degree of sequence identity to a region of the ribosomal assembly protein Noc2 of Aspergillus spp. The cross-reactivity of these regions with AIGAs has been demonstrated in animal models and is considered an example of molecular mimicry. However, EE-IFN-γ only partly restores IFN-γ activity in the plasma of patients with AIGAs, suggesting the presence of other, as yet unidentified, IFN-γ–neutralizing antibodies against different IFN-γ epitopes (Lin et al., 2016).
The clinical manifestations are more variable in subjects with AIGAs than in MSMD patients, including varicella zoster virus reactivation (Chi et al., 2013; Chi et al., 2016; Ku et al., 2020), Talaromyces marneffei infection (Guo et al., 2020), and reactive skin disorders (Browne et al., 2012a; Chan et al., 2013; Jutivorakool et al., 2018; Kampitak et al., 2011; Patel et al., 2005). MSMD patients present an immunological condition resulting in a predominant and selective susceptibility to weakly pathogenic mycobacteria, albeit with a high degree of allelic heterogeneity (Boisson-Dupuis and Bustamante, 2021; Bustamante, 2020; Bustamante et al., 2014; Noma et al., 2022; Rosain et al., 2019). The level of responsiveness to IFN-γ has been proposed to determine the outcome of mycobacterial invasion in patients with disorders of IFN-γR1 or IFN-γR2 (Dupuis et al., 2000). Accordingly, it has been suggested that AIGAs block IFN-γ signaling, based on evaluations of their neutralizing potential in vitro and the poor correlation between plasma AIGA levels and disease severity (Hong et al., 2020; Tham et al., 2016), suggesting a complicated pathogenic effect in patients. The nature of AIGA-related epitope recognition, binding strength, the contribution of the Fc region, and the fluctuations indispensable for blocking IFN-γ activity, either alone or in combination, remain largely unknown. We isolated AIGAs from patients and investigated their physiological effects, with a view to expanding molecular mechanism of action for fine-tuning IFN-γ activity while better delineating the interactive network involved.
Results
Isolation and characterization of pathogenic AIGAs
We investigated the human antibody response to IFN-γ in patients with AIGAs underlying mycobacterial diseases. Three patients presenting with disseminated mycobacterial infection were found to have high plasma IgG titers against recombinant IFN-γ (Fig. S1 A) and to carry at least one HLA-DRB1*16:02–DQB1*05:02 haplotype (Fig. S1 B). After stimulation with IL-12 or IFN-γ in the presence of bacillus Calmette-Guérin (BCG), whole-blood samples from these three patients were found to contain no detectable IFN-γ or IL-12, respectively, suggesting that the AIGAs of these patients had a neutralizing capacity (Fig. S1, C and D). We isolated the AIGA-secreting peripheral memory B cells from these three patients by single-cell sorting based on fluorescently labeled IFN-γ binding to cells of the CD19+CD3−IgG+IgD− subpopulation (Fig. S1 E), for heavy- and light-chain sequence cloning. We obtained 101 mAbs (39 from patient 1, 3 from patient 2, and 59 from patient 3), and 19 clones of these antibodies were found to react with recombinant IFN-γ in IFN-γ–specific ELISAs (Fig. S1 F). Only 10 (5 from patient 1, 3 from patient 2, and 2 from patient 3) of the 19 mAbs and the recombinant AMG811 mAb, a reference anti-human IFN-γ mAb developed for clinical use (Boedigheimer et al., 2017), detected IFN-γ on Western blots, suggesting that the other nine mAbs were conformation sensitive (Fig. S1 G).
Figure S1.
Isolation and generation of pathogenic monoclonal AIGAs from patients underlying mycobacterial diseases. (A) Identification of AIGAs in patient plasma (n = 3) by ELISA. Bar graph of antibody binding to IFN-γ (2 μg/ml) in several dilutions of plasma (100×, 500×, and 2,500×). Each dot corresponds to a donor. (B) HLA-DRB1 and -DQB1 alleles in recruited patients with AIGAs. (C and D) Evaluation of the biological function of AIGAs from three recruited patients. (C) Amount of IFN-γ detected in whole-blood activation assays (NA, BCG, and BCG + 20 ng/ml IL-12) on blood samples from the corresponding donors. Each dot corresponds to a donor. (D) Amount of IL-12p40 detected in whole-blood activation assays (NA, BCG, and BCG + 250 ng/ml IFN-γ) on blood from the corresponding donors. Each dot corresponds to a donor. (E) Isolation of single IFN-γ–specific IgG+CD19+ B cells from patients with mycobacterial disease. Representative flow cytometry plots showing the gating strategy for human IFN-γ–specific B cells from the PBMCs from three patients. Black boxes indicate each successive gate used. Lymphocytes, identified on the basis of forward and side scatter (FSC and SSC), were further analyzed by FSC-A vs. FSC-H comparisons and with 7-aminoactinomycin D staining to eliminate doublets and dead cells. CD19+CD3− cells were then selected, followed by IgG+IgD− cells, to obtain enrichment in specific B cells binding to IFN-γ, which were stained with a FITC-conjugated antibody. IFN-γ+ cells were then sorted from IgG+ subsets. (F) Bar graph showing the reactivity of the 19 monoclonal AIGAs and AMG811 (1 μg/ml) with recombinant human IFN-γ (2 μg/ml), as determined by ELISA. The results are shown as the mean and SD for two independent experiments. (G) Western blot showing the reactivity of 19 monoclonal AIGAs, AMG811, and control IgG (ctrl.; 1 μg/ml) with the recombinant human IFN-γ protein (300 ng/well). The experiments were performed twice, independently. Source data are available for this figure: SourceData FS1.
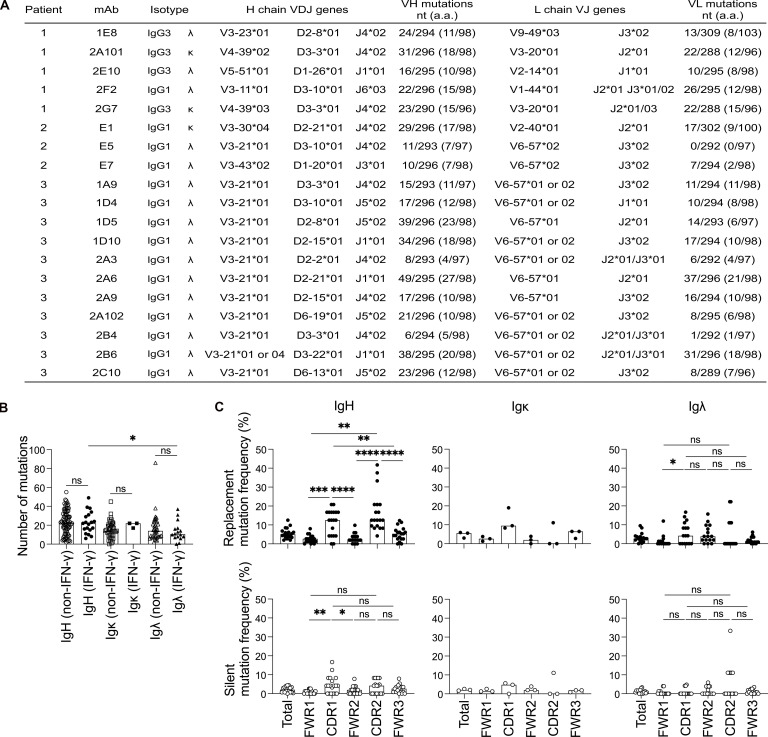
We investigated the genetic features of human AIGAs by analyzing the IgH, Igκ, and Igλ repertoires of isolated mAbs. In total, 15 of the mAbs belonged to the IgG1 subclass; the other 4 belonged to the IgG3 subclass, and λ light-chain usage predominated (16/19; Fig. 1 A). The 2A101 and 2G7 mAbs were encoded by IGHV4-39–D3-3–J4 and IGKV3-20–J2, and were, therefore, considered to correspond to the same clonotype from the same donor (patient 1), but with different somatic hypermutations (SHMs). V gene usage was heavily biased toward the IGHV3-21/IGLV6-57 gene pairing observed in 12 of the 19 mAbs from two individuals (Fig. 1 A), suggesting that IGHV3-21/IGLV6-57 gene usage may be reproducibly induced. In IFN-γ–reactive mAbs, the number of SHMs ranged from 6 to 49 nucleotides in the VH gene segment (mean 28.7 ± 11.5, n = 19) and from 0 to 37 nucleotides in the VL gene segment (mean 20.3 ± 2.8 for Igκ, n =3; 13.4 ± 10.2 for Igλ, n = 16). The number of SHMs was similar between the IFN-γ–reactive and –nonreactive mAbs of patients (Fig. 1 B), consistent with the reported frequency of mutations in human mAbs from healthy donors (Tiller et al., 2007). Substitutions were more frequent than silent mutations in the heavy chain V gene complementarity-determining regions (CDRs), and the frequency of substitutions was higher in these regions than in framework regions, suggesting that the AIGAs had undergone antigen-mediated selection (Fig. 1 C). We therefore concluded that the AIGAs in patients were probably produced by B cells that had undergone affinity maturation, with an Ig-biased usage suggestive of selective dominance among IFN-γ+IgG+ B cells.
Figure 1.
Genetic features of Ig heavy and light chains from the CD19+IgG+ B cells of patients with AIGAs. (A) IgG subclass, isotype, and V, (D), and J usage of Ig heavy and light chains from patient-derived mAbs. (B) Dot plots comparing the SHM (nucleotide mutations) of Ig heavy and light chain (VH, Vκ, and Vλ) genes in non–IFN-γ (n = 82) and IFN-γ–specific (n = 19) CD19+IgG+ B cells. Each dot corresponds to a B cell. The bar represents the median. Mann–Whitney U tests were performed for statistical analysis. *, P < 0.05. (C) The frequency of SHM in VH (n = 19), Vκ (n = 3), and Vλ (n = 16) genes, calculated from the number of replacement (R) and silent (S) nucleotide exchanges per base pair in framework regions (FWRs) and CDRs. Each dot corresponds to a B cell. The bar represents the median. Mann–Whitney U tests were performed for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
Three different groups of high-affinity AIGAs
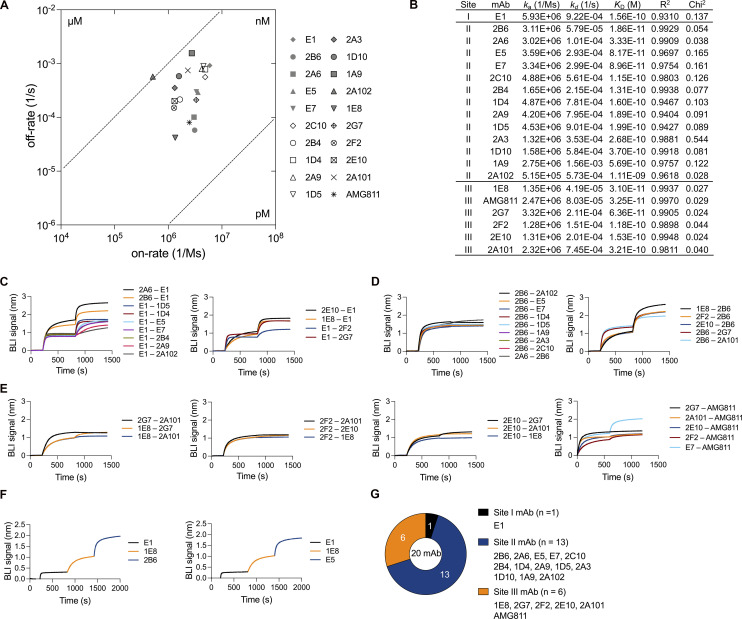
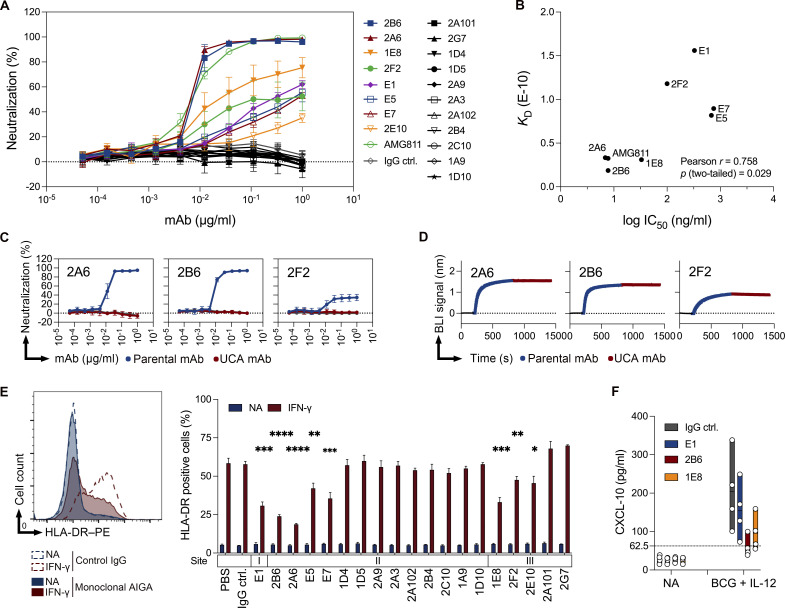
We assessed the binding affinity and epitopes of isolated mAbs and the AMG811 mAb. Biolayer interferometry (BLI) was performed to investigate the kinetics of antibody–antigen interactions. All patient-derived mAbs and the AMG811 mAb had high binding affinities, with dissociation constant (KD) values in the nanomolar range at 1.11 × 10−9 to 1.86 × 10−11 M, and 30% (n = 6) of the isolated antibodies bound to recombinant IFN-γ with a KD < 10−10 M (Fig. 2, A and B; and Fig. S2 A). 18 of the 19 mAbs had similar association rates (1.28–5.93 × 106 1/Ms), whereas binding affinities were more strongly correlated with dissociation rates (Fig. 2 A).
Figure 2.
Binding features of monoclonal AIGAs. (A) Scatter chart of equilibrium KD values, corresponding to the binding affinity, derived from B. KD values were determined from the association (ka) and dissociation (kd) rates of the mAbs (KD = kd/ka). (B) Kinetic values for monoclonal AIGAs and AMG811, which conformed to a 1:1 Langmuir binding model. A χ2 value <3 indicates a good fit of the model to the experimental data. Binding curves are shown in Fig. S2 A. (C–F) Representative graphs showing the in-tandem cross-competition BLI assay for the mAbs and categorizing their binding groups. Biosensor tips were dipped in biotin–IFN-γ (2 μg/ml), then in primary antibody (5 μg/ml), and finally in competing antibody (5 μg/ml). An increase in wavelength (nm) is indicative of binding. The binding readout is depicted in Fig. S2 F. (G) Pie chart showing the three groups of AIGAs (site I, n = 1; II, n = 13; and III, n = 6).
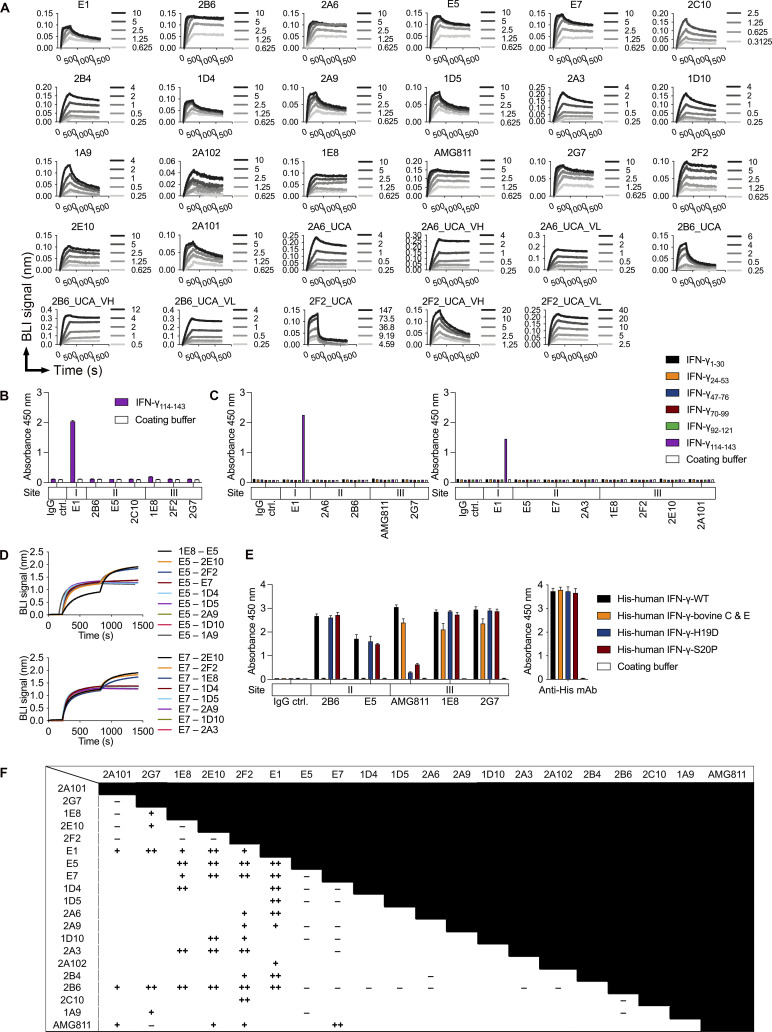
Figure S2.
Kinetic analysis and epitope recognition for monoclonal AIGAs to IFN-γ. (A) Sensorgrams for the binding of 19 monoclonal AIGAs, 9 UCA variants, and AMG811 to the indicated concentrations (nM) of recombinant IFN-γ, determined by BLI. Results for a blank (running buffer without IFN-γ) were subtracted from the results obtained to generate the processed association and dissociation curves. (B) Identification of monoclonal AIGAs binding to synthetic IFN-γ114–143 peptide. The ability of the selected mAbs (1 μg/ml) to bind to a synthetic peptide (100 μg/ml) was assessed by ELISA. The assays were performed twice, independently. The triplicate data are shown as the mean and SD for a single representative experiment. (C) Epitope mapping to assess binding to nonoverlapping human IFN-γ 30-mer synthetic peptides (100 μg/ml) from selected mAbs (1 μg/ml), by ELISA. The results shown were obtained in a single experiment. (D) Validation of IGHV3-21/IGLV6-57 paired mAb binding to IFN-γ through E5 and E7 in an in-tandem cross-competition BLI assay. (E) Protein mapping to assess the binding of selected mAbs (1 µg/ml) to recombinant His-tagged IFN-γ variants (1.5 μg/ml), by ELISA. An antibody against the His tag served as a loading control. The assays were performed twice, independently. The duplicate data are shown as the mean and SD for a single experiment. (F) Summary of in-tandem cross-competition assays for monoclonal AIGAs and AMG811. −, no binding signal detected; +, <0.5 wavelength shift (nm); ++, >0.5 wavelength shift (nm).
We investigated the area on the surface of the IFN-γ molecule targeted by the highly effective mAbs in more detail. The IFN-γ114–143 peptide was chosen for this analysis, principally because the IFN-γ C-terminus was known to react with AIGAs in plasma samples (Lin et al., 2016). Several mAbs recognized IFN-γ on Western blots (Fig. S1 G). While the E1 mAb reacted with IFN-γ114–143 in ELISA (Fig. S2, B and C), the other mAbs were unable to recognize all the six linear peptides covering the full-length human IFN-γ sequence (Fig. S2 C). We then performed in-tandem BLI cross-competition experiments, in which biotinylated IFN-γ (biotin–IFN-γ) was captured on a streptavidin biosensor, with the E1 mAb as a competing antibody. We found that 14 selected mAbs bound biotin–IFN-γ with a wavelength shift (0.3–1.7 nm), with the conservation of E1 mAb binding (Fig. 2 C). E1 was therefore considered to have a single binding epitope (site I) on IFN-γ. Studies of antiviral antibodies have suggested that similar antibody sequence features are required for recognition of the same epitope (Chen et al., 2019). We investigated the recognition of IFN-γ by the related IGHV3-21/IGLV6-57 paired AIGAs, using 2B6 as the competing mAb, due to its very high affinity (Fig. 2 B). The 2B6 mAb clearly outcompeted nine mAbs with the paired IGHV3-21/IGLV6-57 feature (Fig. 2 D). In the reciprocal configuration, using E5 or E7 mAbs as competitors, we found that VH3-21/VL6-57 paired mAbs recognized the region identified as site II (Fig. S2 D). Conversely, five mAbs with different V gene usage and the AMG811 mAb accumulated on biotin–IFN-γ in the presence of the 2B6 or E7 mAbs (Fig. 2, D and E), displaying mutual interference with recognition of the third binding region of IFN-γ, site III (Fig. 2 E). Lower-affinity AIGAs (KD > 10−10 M) did not recognize IFN-γ on Western blots, thereby demonstrating that binding affinity is the determinant for the recognition, by AIGAs of site II of IFN-γ (Figs. 2 B and S1 G). Moreover, we had already identified the helical C and E regions of IFN-γ as possible epitopes of the 2B6 mAb in our patent (US 10,968,274 B2). We therefore constructed a chimeric protein in which the helical C and E regions of human IFN-γ were replaced with bovine homologs to validate this finding (Zuber et al., 2016). The 2B6 and E5 mAbs are completely unable to detect the His-human IFN-γ–bovine C and E protein on ELISA (Fig. S2 E), confirming that the helical C and E regions corresponded to the epitope described as site II. Furthermore, we replaced two amino acid residues of IFNγ (histidine 19 and serine 20) with aspartic acid and proline, respectively (US 7,335,743 B2). The H19D and S20P IFN-γ variants displayed much lower levels of binding to AMG811, suggesting that this region corresponds to the putative site III. These two IFN-γ variants were detected by the other site III mAbs (Fig. S2 E), although the BLI cross-competition assay placed them in the same group as AMG811. The AIGAs were thus generally of high affinity. They recognized three nonoverlapping epitopes on IFN-γ (Figs. 2 G and S2 F), including the previously reported C-terminal region (Lin et al., 2016; Wipasa et al., 2018), the helical C and E regions, and an unidentified region possibly located close to the H19/S20 residues of IFN-γ.
Naive-like unmutated common ancestor (UCA) mAbs against IFN-γ
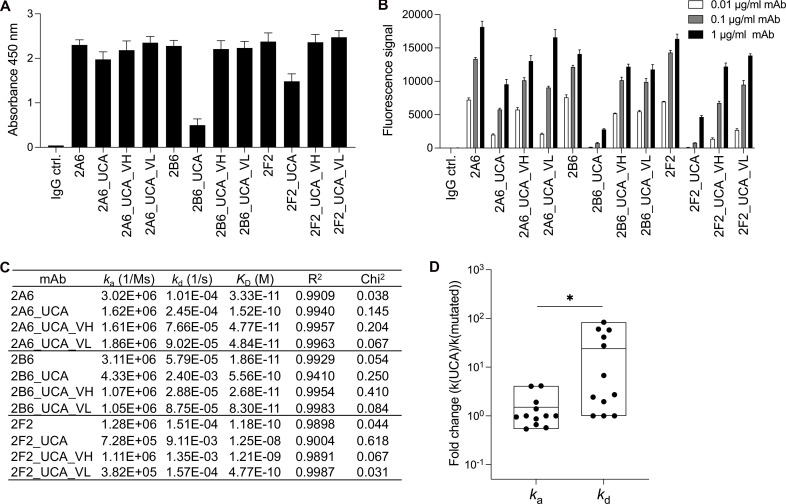
SHM is considered to drive the ability of autoantibodies to break tolerance and recognize the autoantigen (Meffre and Wardemann, 2008). 2Α6 and 2Β6 are highly mutated. They use the IGHV3-21 chain elements with 27 and 20 amino acid substitutions and the IGLV6-57 chain elements with 21 and 18 substitutions, respectively. 2F2 uses an IGHV3-11 chain with 15 amino acid substitutions and an IGLV1-44 chain carrying 12 substitutions. We investigated the effect of SHM on reactivity with IFN-γ by generating UCA variants through SHM reversion in the V gene to the closest matching IMGT germline sequence (Lefranc et al., 2009). We used ELISA and particle-based assays to quantify the binding of UCA mAbs to IFN-γ. The binding signal was weaker, but not abolished (Fig. 3, A and B). The binding level was restored in shuffled variants, in which only the VH or the VL was reverted to the germline sequence (Fig. 3, A and B). The naive-like UCA mAbs of 2A6, 2B6, and 2F2 bound to IFN-γ with affinities of 1.52 × 10−10, 5.56 × 10−10, and 1.25 × 10−8 M, respectively, corresponding to a slight change in affinity (10−1 to 10−2) relative to the parental mAbs (Figs. 3 C and S2 A). In this case, the mutated mAbs had a higher affinity due to lower dissociation rates, with no change in association rates (Fig. 3 D). Nevertheless, naive-like UCA variants bound IFN-γ with nanomolar affinity. Collectively, these data suggest that the AIGA-producing naive B cells are preexisting and already capable of reacting with IFN-γ and undergoing affinity maturation through SHM.
Figure 3.
The contribution of SHM to the ability of antibodies to bind IFN-γ. (A) Bar graph showing ELISA results for reactivity with IFN-γ (2 μg/ml) for three selected highly mutated mAbs (2A6, 2B6, and 2F2; 1 μg/ml), before and after mutation. The control IgG (IgG ctrl.) is a mAb that does not react with IFN-γ. The results are presented as the mean and SD for three independent experiments. (B) The three selected mAbs (2A6, 2B6, and 2F2) were assessed in particle-based assays. The fluorescence signal indicates the ability of the antibody to bind IFN-γ. Two independent runs were performed. The triplicate data are presented as the mean and SD for a single representative experiment. (C) Binding affinity for different UCA variants of AIGAs. The mAbs conformed to a 1:1 Langmuir binding model. A χ2 value <3 indicates a good fit of the model to the experimental data. Binding curves are shown in Fig. S2 A. (D) Fold-changes of ka and kd values between UCA and mutated mAbs (Parental, UCA_VH and UCA_VL) are shown as dots. The results are shown as floating bars from minimum to maximum, with a line indicating the mean. Two-tailed paired Student’s t tests were performed for statistical analysis. *, P < 0.05.
Signal blockade-related neutralization by AIGAs
We evaluated the neutralizing potential of these monoclonal AIGAs for blocking IFN-γ–mediated signaling in vitro in HeLa cells transfected with a IFN-γ activation site (GAS) luciferase reporter plasmid. All the isolated monoclonal AIGAs were tested, and only eight mAbs neutralized IFN-γ signaling, with various potencies, via three binding sites (Figs. 2 G and 4 A). Antibody titration showed that the 2A6 and 2B6 mAbs had strong neutralizing capacities (half-maximal inhibitory concentration [IC50] 6.9 and 7.8 ng/ml, respectively) not inferior to that of a recombinant AMG811 mAb (IC50 7.7 ng/ml; Fig. 4 B). Another six mAbs (E1, E5, E7, 1E8, 2E10, and 2F2) from the three binding groups displayed a moderate and dose-dependent inhibition of the IFN-γ response. Signal blockade was detected principally for high-affinity mAbs for which the strength of binding to IFN-γ was one possible determinant of neutralization. As expected, the inhibitory potential (logIC50) of neutralizing monoclonal AIGAs was positively correlated with binding affinity (KD; Fig. 4 B). Despite the nanomolar scale of binding affinity, 11 mAbs had no inhibitory effect on IFN-γ. The recognition of IFN-γ by AIGAs is not, therefore, sufficient to guarantee their functionality.
Figure 4.
Potential of monoclonal AIGAs to neutralize IFN-γ signaling. (A) In vitro neutralization by 19 monoclonal AIGAs and AMG811 in HeLa GAS reporter cells (2 × 104 cells) treated with 4 ng/ml IFN-γ plus serial threefold dilutions of mAb, beginning at 1 μg/ml, for 20 h at 37°C. Luciferase activity was measured to estimate the percentage of neutralization. The assays were performed at least three times, independently. The results were pooled and are shown as the mean and SD (n = 3–6 per mAb). (B) Correlation between the binding affinity (KD, M) and neutralizing potential (log IC50, ng/ml) of the neutralizing mAbs. 2E10 was not included, because no log IC50 value was available for this mAb. A two-tailed Pearson’s correlation analysis was performed. (C) Assessment of the neutralizing potential of three selected mAbs (2A6, 2B6, and 2F2; serial threefold dilutions and beginning at 1 μg/ml) before and after mutation, in HeLa GAS reporter cells. The results are shown as the mean and SD for three independent experiments. (D) Analysis of competitive binding to sites on IFN-γ for three selected mAbs (2A6, 2B6, and 2F2; 5 μg/ml), before and after mutation, in an in-tandem cross-competition BLI assay. (E) Measurement of HLA-DR expression in THP-1 cells (1 × 105 cells), incubated with and without IFN-γ (20 ng/ml), in the presence of the various mAbs (3 μg/ml), for 24 h at 37°C. Representative histogram (left), as in Fig. S3, and quantitative results (right) for HLA-DR–positive THP-1 cells. The binding sites (I, II, and III) on IFN-γ are indicated above the names of the mAbs. On the right, the results are expressed as the mean and SD for three independent experiments. P values are indicated for comparisons with IgG control (ctrl.) in two-tailed unpaired Student’s t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001. (F) Monoclonal AIGAs inhibit CXCL-10 production. Assessment of CXCL-10 production in a PBMC activation assay involving incubation with the selected monoclonal AIGAs (E1, 2B6, and 1E8; 1 μg/ml) and BCG + IL-12 (10 ng/ml) for 48 h at 37°C. NA, nonactivation. Dashed lines indicate the background signal. Each dot corresponds to a healthy donor of PBMCs. The data are presented as floating bars from minimum to maximum, with a line indicating the mean of four independent experiments.
The correlation between the neutralizing potential and binding affinity of isolated AIGAs and AMG811 mAb suggested that affinity maturation by SHM might be involved in IFN-γ neutralization. We used antibodies with increased SHMs and GAS luciferase reporter assays to test this hypothesis. For three (2A6, 2B6, and 2F2) of the neutralizing AIGAs tested, the UCA versions were unable to neutralize IFN-γ signaling (Fig. 4 C). As previously shown, the UCA mAbs had a high affinity for IFN-γ, but SHM was required to increase affinity further to achieve neutralization. In assessments of epitope-recognition capacity in BLI cross-competition assays, UCA mAbs had an epitope specificity similar to that of the parental mAbs (Fig. 4 D). In particular, AIGAs targeting site II had a similar binding affinity, whereas binding affinity was lower for non-neutralizing clones (KD > 10−10 M; Fig. 2 B) and similar to that of the site II AIGA UCA (Fig. 3 C; 2A6_UCA and 2B6_UCA). However, AIGAs directed against site I or III neutralized IFN-γ with a KD > 10−10 M, suggesting that both affinity and epitope contributed to the neutralization of IFN-γ signaling. These findings indicate that SHM plays a crucial role in generating the higher-affinity antibodies required for neutralization.
IFN-γ is also known as the macrophage-activating factor and has been shown to enhance the maturation of monocytes into mature antigen-presenting cells. We therefore assessed the neutralizing potential of AIGAs by assessing the IFN-γ–responsive expression of HLA-DR on the surface of the human monocyte cell line THP-1. Consistent with the findings of GAS luciferase reporter assays, eight of these mAbs were similarly effective on signal blockade-related neutralization (Figs. 4 E and S3). 2B6 and 2A6 strongly decreased HLA-DR expression on THP-1 cells in the presence of IFN-γ. IL-12 induced robust IFN-γ expression and boosted IFN-γ–responsive CXCL-10 production in PBMCs (Luster and Ravetch, 1987; O’Donnell et al., 1999). For confirmation that AIGAs inhibited the signaling of IFN-γ produced by hematopoietic cells, we measured CXCL-10 production in PBMC activation assays with neutralizing monoclonal AIGAs (E1, 2B6, and 1E8) and BCG + IL-12. These three mAbs inhibited CXCL-10 production (Fig. 4 F). In all three experiments, 2B6 had a greater potential for signal blockade than E1 and 1E8. A higher affinity is, thus, essential for the neutralization of IFN-γ signaling. Monoclonal AIGAs targeting site II strongly neutralized IFN-γ, whereas those targeting site I or III typically displayed moderate neutralization, highlighting the different molecular mechanisms underlying the recognition of different epitopes.
Figure S3.
In vitro neutralizing capacity of monoclonal AIGAs. Analysis of HLA-DR expression in THP-1 cells (1 × 105 cells) in the presence or absence of IFN-γ (20 ng/ml) with the various mAbs (3 μg/ml) for 24 h at 37°C. The assays were performed three times, independently.
Epitope-oriented signal blockade by neutralizing AIGAs
We reasoned that epitope recognition by AIGAs might prevent IFN-γ/IFN-γR binding, thereby exerting constraints on signaling. We tested this hypothesis in an ELISA in which the high-affinity IFN-γR1 protein was immobilized on plates, and the reactivity of biotin–IFN-γ was measured in the presence of various monoclonal AIGAs. Biotin–IFN-γ was detected on the IFN-γR1–coated plates, and binding levels were not modified by increasing the amount of control IgG (Fig. 5 A). In analyses of AIGAs based on their binding sites (I, II, and III), E1 (the only site I mAb) was found to be the most potent of the 15 mAbs tested, dramatically decreasing biotin–IFN-γ reactivity in a dose-dependent manner (Fig. 5 A). By contrast, AIGAs targeting sites II and III had no clear effect on biotin–IFN-γ reactivity (Fig. 5 A). The neutralizing mAbs disrupted cytokine–receptor interaction incompletely, but they only partially inhibited reactivity at a very high concentration (100 nM, corresponding to a mAb/IFN-γ ratio of 10:1). In the reciprocal detection of monoclonal AIGAs, no antibody binding similar to that for the control IgG was detectable for the E1 mAb. By contrast, site II and III mAbs yielded a saturated signal, suggesting that the biotin–IFN-γ was fully occupied by the monoclonal AIGAs (Fig. 5 B).
Figure 5.
Epitope-orientated mechanism of the signal blockade by mAbs. (A and B) Evaluation, by ELISA, of the reactivity between 10 nM biotin–IFN-γ and immobilized 20 nM IFN-γR1 in the presence of mAbs (0, 5, 10, 20, and 100 nM, left to right). The assays were performed at least three times, independently. The results were pooled and are shown as the mean and SD (n = 3–7 per mAb). (A) Bar graph showing the signal for biotin–IFN-γ binding to IFN-γR1 following the addition of the biotin–IFN-γ–mAb mixture. (B) Bar graph showing the signal for the binding of the mAb to IFN-γR1 via biotin–IFN-γ. (C and D) IFN-γ–mediated binding of mAbs (40 nM) to THP-1 cells (1 × 105 cells). The amount of antibody binding to the cell was determined by flow cytometry with anti-human IgG–PE antibodies. Representative histogram (C) and quantitative results (D) for the mean fluorescence intensity (MFI) of IgG–PE from THP-1 cells in the presence and absence of IFN-γ (20 nM). The binding sites (I, II, and III) on IFN-γ are indicated above the mAbs. The results are shown as the mean and SD for three independent experiments. (E) Labeling of IFN-γR1 (purple) and IFN-γR2 (green) in the plasma membrane (PM) of live cells with Rho-11– (red) and AT643-conjugated (blue) anti-GFP nanobodies, for the assessment of IFN-γR1/IFN-γR2 dimerization by single molecule cotracking (magenta circle). Heterodimerization (top) was detected as the orthogonal labeling of IFN-γR1 and IFN-γR2 with mXFPm/MIRho11 and mXFPe/ENAT643, respectively. IFN-γR2 homodimerization (bottom) was detected as the labeling of mXFPe-IFN-γR2 with a mixture of ENRho11 and ENAT643. EN, enhancer; MI, minimizer. (F) Quantification of the heterodimerization of IFN-γR1 and IFN-γR2 with anti-GFP nanobodies labeled with Rho11 and AT643. Relative cotracking of Rho11–IFN-γR1 and AT643–IFN-γR2 in the presence of IFN-γ (10 nM) and mAb (20 nM). The data presented are means ± SEM; IgG Ctrl. (n = 20), E1 (n = 18), 2B6 (n = 19), 2A102 (n = 16), 1E8 (n = 19), 2G7 (n = 19). Each data point corresponds to a cell. (G) Quantification of the homodimerization of IFN-γR2. The data presented are means ± SEM; IgG Ctrl. (n = 20), E1 (n = 19), 2B6 (n = 20), 2A102 (n = 18), 1E8 (n = 22), 2G7 (n = 19). Each data point represents a cell. The P values in panels F and G were calculated in two-tailed unpaired Student’s t tests. **, P < 0.01; ****, P < 0.001. The binding sites (I, II, and III) on IFN-γ are indicated above the mAbs. (H and I) Evaluation of the synergistic effect on neutralization of mAbs (serial threefold dilutions and beginning at 1 μg/ml in total) based on binding sites (I, II, and III) and neutralizing capacity in HeLa GAS reporter cells (2 × 104 cells) within 4 ng/ml IFN-γ. In vitro neutralizing potential of groups of neutralizing mAbs (H) Site I, E1; site II, E5; site III, 1E8 (I); site I: E1; site II: 2B6; site III: 1E8 (J). In vitro neutralizing potential of groups of non-neutralizing mAbs. Site II: 2C10 and 1D5; site III: 2G7 and 2A101. 2A6 is a neutralizing mAb. (K) In vitro neutralizing potential of a group of neutralizing and non-neutralizing mAbs. Site I: E1 with neutralization; site II: 2C10, and site III: 2G7 without neutralization. (L and M) In vitro dose-dependent neutralizing potential of neutralizing mAbs (E1 or 2B6; serial threefold dilutions and beginning at 1 μg/ml) in the presence of a non-neutralizing mAb (2G7; serial threefold dilutions and beginning at 1, 3, or 9 μg/ml) with one, three, or ninefold dilutions. Control IgG, IgG ctrl. The results in H–M are shown as the mean and SD for three independent experiments. Two-tailed paired Student’s t tests were used to compare 1 μg/ml mAb treatments in H and K–M. ns, non-significant; *, P < 0.05; ***, P < 0.005.
We then extended the concept to a cell-based assay, in which the reactivity of AIGAs with THP-1 cells expressing the native IFN-γR, via IFN-γ, was assessed by flow cytometry. We chose nine monoclonal AIGAs from the three binding groups at random, regardless of their neutralizing potential. All eight site II or III monoclonal AIGAs were detected and were presumably anchored at the cell surface via IFN-γ (Fig. 5, C and D). By contrast, the absence of a fluorescent signal following the addition of E1 suggests that the IFN-γ/E1 mAb complex cannot attach to IFN-γR–expressing cells (Fig. 5, C and D), consistent with the ELISA data (Fig. 5, A and B). Thus, the E1 mAb disrupts the interaction of IFN-γ with the high-affinity IFN-γR1 subunit of the receptor, thereby efficiently preventing cell activation by IFN-γ.
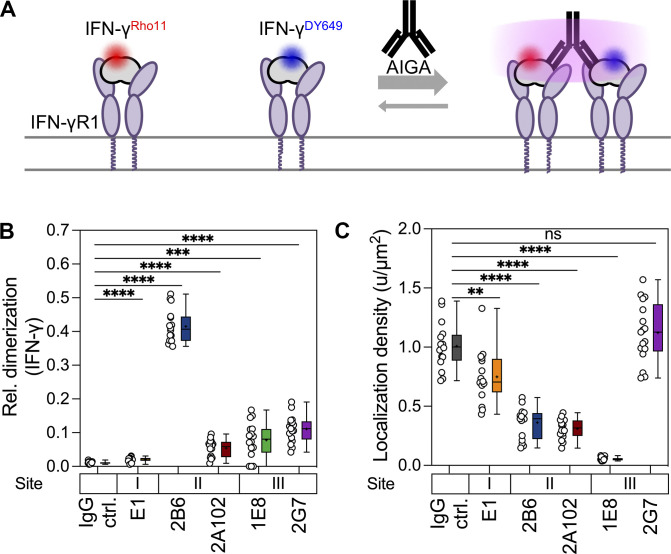
Neutralizing AIGAs directed against site II or III didn’t affect the binding of IFN-γ to the IFN-γR (Fig. 5 C). This indicates that, in addition to disrupting IFN-γ binding to IFN-γR, other mechanisms underly the interruption of IFN-γ signaling by AIGAs. IFN-γ is a dimeric ligand that binds with high affinity to the IFN-γR1 subunit, triggering its dimerization, leading to the recruitment of the IFN-γR2 subunit, which is strictly required for signaling (Greenlund et al., 1993; Mendoza et al., 2019). We therefore hypothesized that neutralizing AIGAs against sites II and III might prevent IFN-γR2 from coming into close contact with the IFN-γ/IFN-γR1 complex at the cell surface. We performed two-color single-molecule cotracking experiments by total internal reflection fluorescence (TIRF) microscopy to quantify the IFN-γ–stimulated dimerization of IFN-γR1 and IFN-γR2 in the plasma membrane (Mendoza et al., 2019; Fig. 5 E). IFN-γ promoted the heterodimerization of IFN-γR1 and IFN-γR2 and the homodimerization of IFN-γR2 in the presence of control IgG (Fig. 5, F and G). Following the addition of monoclonal AIGAs and IFN-γ, much lower levels of IFN-γR1 and IFN-γR2 heterodimerization were observed in the presence of the 2B6 and 1E8 mAbs (Fig. 5 F). Conversely, the levels of IFN-γ–induced IFN-γR1 and IFN-γR2 heterodimers were not decreased by the two non-neutralizing mAbs, 2A102 and 2G7, in these assays (Fig. 5 F). Likewise, IFN-γR2 homodimerization was strongly decreased by monoclonal AIGAs against site II or III (Fig. 5 G). Together, these data demonstrate that the binding of 2B6 and 1E8 (sites II and III, respectively) to IFN-γ interferes with the recruitment of IFN-γR2 to the signaling complex, resulting in signal blockade. Consistent with this hypothesis, IFN-γ binding to the cell surface was largely uncompromised in the presence of site II and III mAbs, which induced IFN-γ cross-linking (Fig. S4). Interestingly, E1 decreased IFN-γ binding to the cell surface only slightly, consistent with its weak inhibitory function (Fig. 4 A), potentially due to ineffective competition with the cooperative interaction of IFN-γR1 and IFN-γR2 at the cell surface (Fig. 5, F and G). These modes of binding define distinct AIGA-modulated IFN-γ signaling profiles, probably resulting from differences in the orchestration of the AIGA/IFN-γ/IFN-γR complex.
Figure S4.
Dimerization and cell surface receptor binding of IFN-γ in the presence of monoclonal AIGAs. (A) Rho11-labeled IFN-γ (red; 5 nM) and DY649-labeled IFN-γ (blue; 5 nM) were used to assess the binding to the cell surface receptor and potential dimerization induced by monoclonal AIGAs (20 nM). (B) Quantification of ligand homodimerization with Rho11- and DY649-conjugated IFN-γ. Relative tracking of IFN-γRho11 and IFN-γDY649 in the presence of monoclonal AIGAs, as determined by single-molecule cotracking. (C) Quantification of cell surface-bound IFN-γ density in the presence of monoclonal AIGAs by single-molecule localization analysis. The data shown are the mean ± SEM. IgG ctrl (n = 16), E1 (n = 16), 2B6 (n = 16), 2A102 (n = 16), 1E8 (n = 16), 2G7 (n = 17); each data point represents a cell. The binding sites (I, II, and III) on IFN-γ are indicated above the monoclonal AIGAs. The P values in B and C were calculated in unpaired two-tailed Student’s t tests. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Non-neutralizing AIGAs do not exhibit synergistic effect on modulating IFN-γ signaling
We identified three neutralization-sensitive binding sites on IFN-γ, and a robust neutralization of IFN-γ signaling was observed in the plasma of most patients (Aoki et al., 2018; Chi et al., 2013; Guo et al., 2020). We evaluated the neutralizing potential of polyclonal antibody responses to IFN-γ by designing cocktails of mAbs based on a matrix of neutralizing ability and binding sites in equimolar ratios, resulting in three groups: (a) a group of neutralizing mAbs, (b) a group of non-neutralizing mAbs, and (c) a group of mixed mAbs. We measured the neutralizing potential of these cocktails in the GAS luciferase reporter assay. Combinations of two or three moderately neutralizing mAbs displayed remarkable synergy in terms of signal blockade, with the use of the E1 + E5 + 1E8 combination of mAbs strengthening the neutralization of IFN-γ signaling, with full neutralization achieved at high concentrations of these mAbs (Fig. 5 H). We then replaced E5 with 2B6 in a similar setting. 2B6 is a fully neutralizing mAb that plays the dominant role in combinations of two or three mAbs (Fig. 5 I). In a reciprocal setting, a group of non-neutralizing mAbs clearly had no qualitative effect on neutralization (Fig. 5 J). In addition, a group of mixed mAbs inhibited IFN-γ no more strongly than the individual mAbs, whether used at an equimolar ratio (Fig. 5 K) or with the gradual addition of the non-neutralizing mAb, 2G7 (Fig. 5, L and M). Overall, these results suggest that the potential to neutralize IFN-γ signaling can be attributed to neutralizing mAbs. Non-neutralizing mAbs did not appear to act synergistically, even in the presence of negligible antagonism due to steric hindrance.
The role of immunocomplexes in neutralizing IFN-γ signaling
Theoretically, dimeric IFN-γ could form high molecular weight immunocomplexes with a single monoclonal AIGA with bivalent interactions. We therefore investigated immunocomplex formation in equimolar mixtures of mAb and dimeric IFN-γ, by performing size-exclusion chromatography/ultra-high-performance liquid chromatography (SEC-UHPLC). Site II monoclonal AIGAs had a much greater tendency to generate immunocomplexes with IFN-γ than site I or III monoclonal AIGAs (Fig. 6 A). In particular, 2A6 and 2B6 formed higher-molecular weight immunocomplexes (>1,000 kD) with IFN-γ (Fig. 6, A and B). These findings suggest that the recognition of epitopes at site II of IFN-γ leads to immunocomplex generation.
Figure 6.
Immunocomplex formation by the monoclonal AIGAs. (A) SEC profiles of samples (40 μl) containing a single antibody (3 μΜ) with or without IFN-γ (3 μΜ). Blue traces, mAb alone; red lines, samples with both mAb and IFN-γ; AU, absorbance unit. The assays were performed twice independently. (B) The dose dependence of immunocomplex formation was investigated with 1:0, 1:0.5, 1:1, and 1:2 ratios of site II mAb (2B6 and 2C10; 3 μM) to IFN-γ (0, 1.5, 3 or 6 μM) in SEC-UHPLC analysis. The assays were performed twice independently. (C) In vitro Fab-based neutralization in HeLa GAS reporter cells. Monovalent Fab fragments were generated by digesting the IgGs with the Pierce Fab micropreparation kit, according to the manufacturer’s instructions. HeLa GAS reporter cells (2 × 104 cells) were stimulated with 118 pM (4 ng/ml) IFN-γ in the presence of serial threefold dilutions of Fab, beginning at 120 nM. The full-length antibody was used as a control, under serial threefold dilutions, beginning at 6.7 nM. The results are shown as the mean and SD for two independent experiments. The binding sites (I, II, and III) on IFN-γ are indicated next to the mAbs and Fabs.
The 2B6 site II mAb blocked IFN-γ signaling by preventing IFN-γ receptor assembly (Fig. 5, F and G), but we wondered whether the immunocomplex played a role in signal blockade. In the HeLa GAS reporter assay, the monovalent Fab fragments of 2B6, which are unable to form immunocomplexes with IFN-γ, had qualitative effects similar to those of the corresponding antibody, but weaker (Fig. 6 C). The blockade of IFN-γ signaling was, therefore, independent of antibody bivalency and the formation of high-molecular weight immunocomplexes. Further, 1E8 had neutralizing activity but did not form immunocomplexes (Fig. 6, A and C). We can, therefore, conclude that immunocomplex formation is not essential for neutralization and interference with IFN-γ receptor assembly.
Fc-dependent role of monoclonal AIGAs
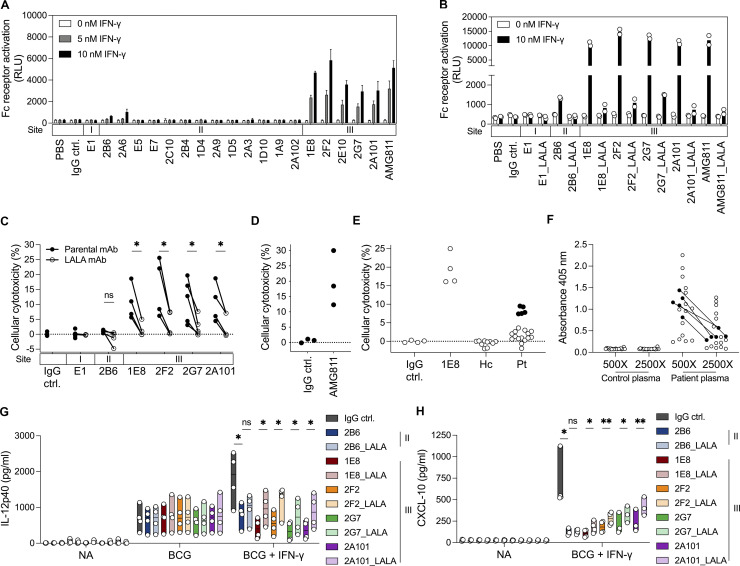
Monoclonal AIGAs were attached to IFN-γR–expressing cells via targeting site II or III on IFN-γ (Fig. 5 C). The Fc region of the attached AIGAs may, therefore, have biological activity through recognition of the FcγR on effector cells (Clynes and Ravetch, 1995; Nimmerjahn and Ravetch, 2005). We investigated the contribution of the Fc region of AIGAs in a T cell activation bioassay system (Parekh et al., 2012). We engineered Jurkat T cells stably expressing FcγRΙΙIa, the low-affinity receptor for monomeric IgG, and the luciferase reporter gene under the control of nuclear factor of activated T-cells–response element (NFAT-RE). These cells were used as effector cells and were cocultured with IFN-γR–expressing THP-1 cells, used as target cells. Individual mAbs were mixed with IFN-γ in the coculture system, and luciferin production was assessed, as a means of estimating FcγRIIIa activation. We tested all the isolated mAbs and AMG811. Site III mAbs boosted luciferin production in a manner dependent on the dose of IFN-γ (Fig. 7 A). The luciferase activity of the effector cells was barely detectable following conditioning with mAbs against site I or II, regardless of the observed anchoring of site II mAbs on the cell surface in the presence of IFN-γ (Fig. 5 C). However, these site II mAbs were unable to activate luciferase strongly via FcγRIIIa (Fig. 7 A), possibly due to steric hindrance rendering the Fc region inaccessible to the Fcγ receptor (Renner et al., 2018). We validated the Fc region of AIGAs in activating FcγR signaling. We introduced L234A/L235A (LALA) mutations into the Fc region of AIGAs, which decreases the strength of Fc interaction with FcγR (Hezareh et al., 2001; Jefferis et al., 1990; Wilkinson et al., 2021). The LALA variants of site III mAbs produced only small amounts of luciferin (Fig. 7 B), suggestive of abolition of the Fc–FcγR interaction.
Figure 7.
Monoclonal AIGAs selectively facilitate cellular cytotoxicity. (A) Fc receptor activation indicated by the bar graph showing luciferase activity in 5 × 105 Jurkat FcγRIIIa–NFAT-RE reporter cells cocultured with 1 × 105 THP-1 cells in the presence of IFN-γ (0, 5, and 10 nM) and mAb (10 nM) for 6 h at 37°C. The results are shown as the mean and SD for three independent experiments. (B) Fc receptor activation status for the selected LALA variants of the mAbs (10 nM) and IFN-γ (0 and 10 nM) in 5 × 105 Jurkat FcγRIIIa–NFAT-RE reporter cells cocultured with 1 × 105 THP-1 cells for 6 h at 37°C. The dot points are from two independent experiments, and the bar graph is shown as mean. (C) Cytotoxicity was determined with 5 × 104 NK cells as effector cells (E) and 5 × 103 monocytes as target cells (T; E:T = 10:1), incubated with 10 nM IFN-γ and 10 nM mAb for 8 h at 37°C. Each dot corresponds to a healthy donor of primary cells. The assays were performed at least three times independently and the results were pooled (n = 3–6 per mAb). P values were calculated in two-tailed paired Student’s t tests. *, P < 0.05. The binding sites (I, II, and III) on IFN-γ are indicated above the mAbs. (D and E) Cellular cytotoxicity of AMG811 determined with 1 × 105 NK cells as effector cells (E) and 5 × 103 monocytes as target cells (T; E:T = 20:1), incubated with 10 nM IFN-γ and 10 nM mAb for 8 h at 37°C. Each dot corresponds to a healthy donor of primary cells, in three independent experiments. IgG control, IgG ctrl. (E) Plasma (2%) from individuals with (n = 20) or without (n = 10) AIGAs was assayed for cytotoxicity mediated via IFN-γ (E:T = 20:1) for 8 h at 37°C. Each dot corresponds to a healthy donor of primary cells. Pooled data from four independent experiments are shown. ctrl, control; Hc, healthy control; Pt, patient with AIGAs. 1E8 mAb was used as a positive control. (F) Identification of AIGAs from selected donors (n = 20) for cytotoxicity assays by ELISA. Dot plot showing the ability of antibodies to bind to IFN-γ (2 μg/ml) in serial dilutions of plasma (500× and 2,500×). Black dots, five donor plasma samples displaying ADCC in the cytotoxicity assay. (G) ELISA quantification of IL-12p40 production by PBMCs for the NA, BCG, and BCG + IFN-γ (25 ng/ml) treatments in the presence of 1 μg/ml mAb for 48 h at 37°C. The binding sites (II and III) on IFN-γ are indicated next to the monoclonal AIGAs. Each dot corresponds to a healthy donor of PBMCs. The data are shown as floating bars from minimum to maximum, with a line indicating the median of four independent experiments. (H) ELISA quantification of CXCL-10 production by PBMCs for the NA and BCG + IFN-γ (25 ng/ml) treatments, in the presence of 1 μg/ml mAb for 48 h at 37°C. The assays were performed at least three times, independently. Each dot corresponds to a healthy donor of PBMCs. The results were pooled and are shown as floating bars from minimum to maximum, with a line indicating the median (n = 3–4 per mAb). In G and H, two-tailed paired Student’s t tests were used for statistical analysis. *, P < 0.05; **, P < 0.01.
Most of the AIGAs in patients and most of the mAbs isolated in this study belong to the IgG1 subclass, which is a potent activator of antibody-dependent cellular cytotoxicity (ADCC; Fig. 1 A; Browne et al., 2012a). We investigated whether enhanced antibody-mediated cell bridging by site III monoclonal AIGAs contributed to ADCC. We performed cytotoxicity assays with isolated NK cells expressing FcγRIIIa as effector cells and autologous monocytes expressing IFN-γR as target cells, in which we assessed the cell lysis level in the presence of equimolar amounts of mAb and IFN-γ. Treatment with site III mAbs resulted in marked cell lysis, with mean values of 10.54% for 1E8 (range 5.67–18.65%), 15.56% for 2F2 (range 6.17–25.60%), 11.26% for 2G7 (range 3.09–19.70%), and 10.43% for 2A101 (range 4.45–18.74%). While the AMG811 recognizes site III on IFN-γ, we also found that AMG811 exhibited high cytotoxicity, with a mean value of 20.23% (range 12.30–30.00%; Fig. 7 D). Cytotoxicity was Fc dependent, as significantly fewer lysed cells were observed with LALA variants of the site III mAbs than with the parental mAbs. Almost no cell lysis was detected in cells treated with 2B6 (0.94%, range 0.35–1.47%) or E1 (0.03%, range −0.54–1.91%; Fig. 7 C). We then examined the possibility of ADCC being mediated by AIGAs from patient plasma ex vivo. We found that plasma samples from 5 of the 20 patients with AIGAs tested displayed cytotoxicity, with 7.4–9.5% of cells lysed, whereas the positive control (1E8) gave a mean of 19.3% cell lysis (Fig. 7, E and F). Consistent with these findings, the 1E8 mAb was obtained from one of these five patients. Only 25% (5/20) of the subjects presented moderate cell lysis through ADCC, and plasma from the other patients was not cytotoxic, potentially reflecting possible differences in AIGA composition between these two groups of patients (Fig. 7 E). Collectively, AIGAs directed against site III on IFN-γ can activate effector cells and mediate ADCC targeting IFN-γR–expressing cells.
We then investigated the impact of ADCC on hematopoietic cells (e.g., monocytes/macrophages) in terms of their IFN-γ responsiveness, which was assessed by measuring IL-12p40 and CXCL-10 levels in an ELISA on PBMCs stimulated with BCG in the presence of IFN-γ and mAbs. Simulation with BCG + IFN-γ induced the production of larger amounts of IL-12p40 by the monocytes than stimulation with BCG alone (Fig. 7 G). In particular, site III mAbs inhibited the production of IL-12p40 more strongly than 2B6, which specifically abolished signaling (Fig. 7 G), and 2G7 and 2A101 were found to be non-neutralizing in reporter-based functional assays in HeLa cells (Fig. 4 A) but markedly decreased the levels of IL-12p40 and CXCL-10 in PBMC activation assays (Fig. 7, G and H), demonstrating that the response was entirely dependent on the effector cells and their interaction with antibody. Treatment with LALA variants of AIGAs had a significantly weaker inhibitory effect on IL-12p40 and CXCL-10 production than the parental site III mAbs. NK cells with abundant FcγIIIa are considered an effective cytolytic leukocyte subset of PBMCs. LALA variants of anti–HIV-1 antibodies have been shown to decrease antibody binding to FcγRIIa and FcγRΙΙΙa dramatically, but to have no effect on binding to FcγRI (Hessell et al., 2007; Hezareh et al., 2001). The site III mAbs studied here tended to produce smaller amounts of IL-12p40 after incubation with BCG + IFN-γ than after incubation with BCG alone, due to the FcγRIIIa-mediated killing of monocytes by AIGAs. These data highlight a potential role of Fc–FcγR interactions mediated by pathogenic AIGAs not coupled to the Fab-mediated blockade of IFN-γ downstream signaling. The biased position on site III of IFN-γ enables the mAbs to mediate ADCC, thereby decreasing any function by IFN-γR–expressing cells, such as IL-12 production.
Discussion
AIGAs are the underlying factor predisposing patients to infectious diseases by targeting endogenous IFN-γ. Nevertheless, the molecular mechanism of AIGA pathogenesis and the contribution of these antibodies to the immune system have been little studied. We isolated AIGAs from patients, and we provide here a detailed characterization of their molecular features and pathogenic effects (Fig. S5). We showed that SHMs contribute to increasing binding affinity of AIGAs to IFN-γ in neutralizing IFN-γ signaling. A pre-existing ability to bind IFN-γ was demonstrated for near-germline antibodies, independently of signal blockade. Epitope binding analyses classified these AIGAs into three separate groups. These mAbs neutralize IFN-γ signaling in several different ways—by preventing receptor binding or interfering with receptor assembly—and the mode of action used depends on epitope group. In addition to their ability to blockade IFN-γ signaling, AIGAs may anchor themselves to cells via IFN-γ/IFN-γR interactions, inducing ADCC to kill IFN-γR expressing cells through Fc-dependent activity, thereby decreasing IL-12p40 and CXCL-10 production. Collectively, these molecular features and functional dissections of AIGAs provide important information about the production of these autoantibodies and the mechanism by which they impair host immunity, thereby explaining their association with susceptibility to diverse infectious diseases and paving the way for the development of therapeutic or disease prevention strategies.
Figure S5.
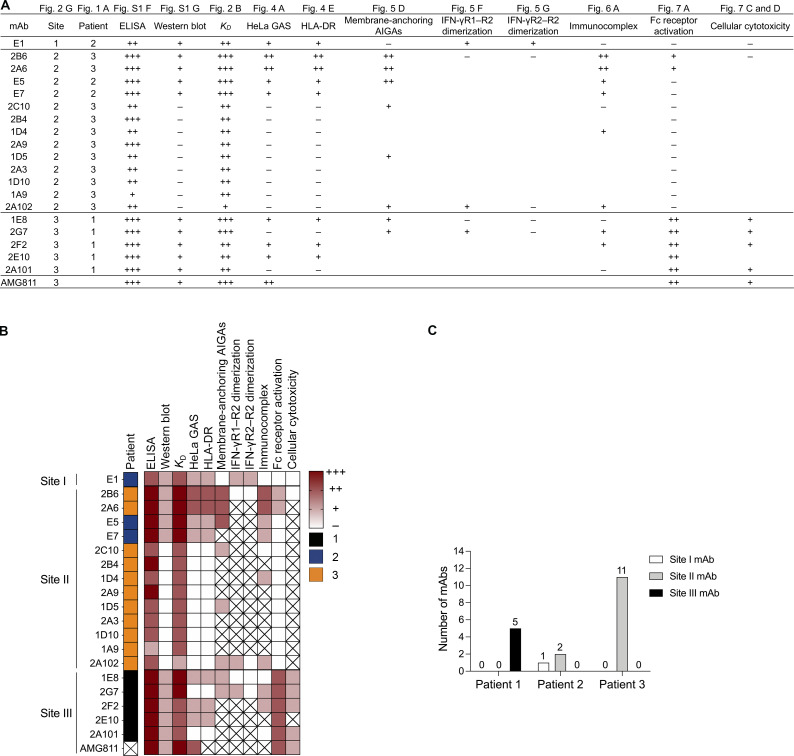
Summary information for the 19 monoclonal AIGAs and AMG811. (A and B) Patient distribution and binding characteristics (epitope group, ELISA, Western blot, binding affinity, immunocomplex) and functional impacts (HeLa GAS reporter, HLA-DR expression, IFN-γR1–R2 and IFN-γR2–R2 dimerization, Fc receptor activation, cellular cytotoxicity) of the 20 mAbs indicated in table (A) and heatmap (B). +++, ++, +, − indicate ability, strength, or potency; 1, 2, 3 indicate patient category; X, an absence of measurement in assays. (C) Bar graph for the distribution of the 19 monoclonal AIGAs on the basis of patients and binding sites.
We found that SHM was involved in autoantibody development and crucial for the neutralizing potential of AIGAs. The SHM levels of the mAbs isolated, which were similar to those reported elsewhere for other mAbs, suggest a role in classical T cell–dependent maturation. We studied naive-like UCA mAbs and found that (a) they bound IFN-γ with high affinity (KD, 10−8∼10−10 M), indicating that B cell receptors, with preferential IGHV3-21/IGLV6-57 usage on naive B cells, could recognize IFN-γ or relevant structures; (b) SHM increased the affinity of AIGAs for IFN-γ; and (c) SHM was required for signal blockade rather than for specificity. However, near-germline antibodies have been reported to bind to and neutralize pathogens such as respiratory syncytial virus and SARS-CoV-2 (Goodwin et al., 2018; Kreer et al., 2020). Germline antibody genes are thus considered to constitute a selective advantage, as they make it possible to generate antibodies immediately and to fight evolving infectious pathogens. The biological role of high-affinity germline-like antibodies against IFN-γ remains unclear. Pathogenic AIGAs may be developed from high-affinity germline-like antibodies, accelerating tolerance-breaking mechanisms.
AIGAs recognized three epitopes and impeded IFN-γ signaling by preventing IFN-γ binding and disrupting the active IFN-γR complex, in a process that was unexpected for anticytokine autoantibodies. The E1 mAb targeted the C-terminus of IFN-γ (IFN-γ114–143), which has been reported to carry a major epitope for neutralization by AIGAs in plasma from patients (Lin et al., 2016; Wipasa et al., 2018). The binding of E1 to the C-terminus of IFN-γ resulted in a signal blockade, through prevention of the binding of the cytokine to IFN-γR1. Consistently, anti-human IFN-γ C-terminus antibodies raised in mice block IFN-γ signaling (Seelig et al., 1988), and the C-terminal region has been identified as an interface between IFN-γ and IFN-γR1 (Lundell and Narula, 1994; Walter et al., 1995). Furthermore, the helical C and E regions, or a possible region close to His19/Ser20 in IFN-γ, are identified by two other groups (site II and III) of mAbs that neutralize IFN-γ signaling more potently without disrupting the binding of IFN-γ to its cell surface receptor. Instead, these antibodies dock with IFN-γ, abolishing the dimerization of IFN-γR1 and IFN-γR2 and preventing signaling activation. IFN-γR1, a high-affinity receptor for IFN-γ, dimerizes upon binding to homodimeric IFN-γ, further stabilizing the binding of the accessory subunit IFN-γR2. However, the second copy of IFN-γR2 is redundant, and therefore unnecessary for complete IFN-γ signaling (Mendoza et al., 2019). A total loss of contact between IFN-γR2 and IFN-γR1, disabling IFN-γ signaling, has been reported in MSMD patients with T168N mutations of the IFN-γR2 gene (Vogt et al., 2005), and in biophysical assays with engineered IFN-γ (Mendoza et al., 2019). The genetic evidence, biophysical data, and our findings all indicate that the disruption of IFN-γ–induced IFN-γR assembly can cause IFN-γ dysfunction. The identification of neutralizing antibodies targeting epitopes outside the C-terminal region of IFN-γ provides an explanation for our previous observation that EE-IFN-γ results in partial restoration in only some AIGA patients, with no effect in others. All the site III mAbs were obtained from patient 1, and 11 of the 13 site II mAbs came from patient 3. Patient 2 had site I and II mAbs (Fig. S5). EE-IFN-γ therefore appears to be more functional in some individuals (e.g., patient 2) with a predominant AIGA response to C-terminal IFN-γ, than in others with a different response pattern (e.g., patients 1 and 3).
We further demonstrated that reconstituted mixtures with non-neutralizing mAbs could not block IFN-γ signaling, possibly due to insufficient binding affinity. Conversely, combinations of neutralizing monoclonal AIGAs with extremely high affinities had synergistic effects (Fig. 5, H and I). The neutralization potential of the AIGAs in plasma from patients is, therefore, determined by the presence of high-affinity neutralizing antibodies rather than the concentration of AIGAs, which include both neutralizing and non-neutralizing antibodies directed against IFN-γ. This finding provides a plausible explanation for AIGA concentration not reflecting disease severity (Hong et al., 2020), which may instead depend on the fraction of neutralizing antibodies. Regardless of antibody titer, the neutralizing potential may, therefore, be a better biomarker for predicting the severity of disease in patients with AIGAs. By contrast, in patients with pulmonary alveolar proteinosis and autoantibodies against GM-CSF, GM-CSF is cleared via cooperative immunocomplexes involving several non-neutralizing antibodies (Piccoli et al., 2015). In this case, the concentration of autoantibodies must be high to ensure adequate immunocomplex formation with GM-CSF, to reduce the activity of this factor and lead to the clinical onset of pulmonary alveolar proteinosis (Uchida et al., 2004). AIGAs and anti–GM-CSF autoantibodies impede the signaling of cognate cytokines via different mechanisms. A characterization of the mode of action of these antibodies is important to improve the diagnosis and treatment of these diseases, and may also be crucial for other anticytokine autoantibody diseases, such as those involving anti-type I IFN autoantibodies (Bastard et al., 2020).
Our findings demonstrate that, in addition to their blockade of IFN-γ signaling, AIGAs may be pathogenic due to their ability to activate Fc-mediated effector functions. The loss of IFN-γ–responsive cells with site III monoclonal AIGAs was mostly accompanied by a drop of ISG expression via ADCC, which can be indicative of pathogenic AIGAs. The relative contribution of Fc-mediated responses in patients remain uncertain. Macrophages are the principal cells involved in the response to IFN-γ and are also the major source of IL-12/IL-23. We speculate that macrophages may be subject to ADCC during infection, leading to an impairment of IL-12/IL-23 production in patients carrying AIGAs directed against site III epitope. It has been suggested that IL-12/IL-23 act partly through an IFN-γ–independent pathway in immunity to Salmonella in humans (MacLennan et al., 2004). Interestingly, almost one third of patients with AIGAs underlying mycobacterial infection had a history of coinfection with Salmonella spp. (Chi et al., 2016), which is common in patients with IL-12–related defects, but rarely seen in cases of IFN-γR deficiency (Bustamante et al., 2014; MacLennan et al., 2004). Moreover, either cutaneous involvement in mycobacterial infection or Sweet’s syndrome/neutrophilic dermatosis (Chan et al., 2013; Chaowattanapanit et al., 2016; Jutivorakool et al., 2018; Kampitak et al., 2011) is increasingly regarded as one of many clinical features (Shih et al., 2021) observed in patients with AIGAs but not in those with MSMD. It may be worth investigating whether the AIGA-dependent killing of macrophages by ADCC and impairment of IL-12/IL-23 can cause salmonellosis or dermatosis in some patients with AIGAs predominantly directed against site III. The neutralization of IFN-γ signaling and Fc-mediated cellular cytotoxicity due to AIGAs directed against various epitopes may contribute to the illness and to particular manifestations of AIGA-mediated diseases.
Both site II and III monoclonal AIGAs could interfere with assembly of the IFN-γ receptor complex. However, the precise manner in which these AIGAs cause steric hindrance in receptor assembly is unknown. Additional studies providing structural insight are required to elucidate the precise epitopes and spatial conformations involved in the interruption of receptor assembly. Structural investigations might also provide an explanation for the biased Fc-dependent activity of AIGAs, which may reflect differences in Fc receptor accessibility. Such a situation has already been described for two potent and neutralizing antibodies against dengue virus occluding largely overlapping epitopes but with different abilities to bind to the Fc receptor. (Renner et al., 2018). Site II monoclonal AIGAs may have bound to IFN-γ and presented a flat angle on IFN-γR–expressing cells, thereby restricting the FcγR interaction. Furthermore, an impairment of the cellular response to IFN-γ by AIGAs has been implicated in assays of the neutralization of STAT1 phosphorylation (Aoki et al., 2018; Browne et al., 2012a; Lin et al., 2016), which provide no information about the Fc-dependent activity of AIGAs. However, measurement of the Fc-mediated response of AIGAs may be required for full elucidation of their pathogenic role. Moreover, our previous studies have indicated that a modified IFN-γ (EE-IFN-γ) can restore IFN-γ activity by escaping autoantibodies binding to the C-terminus of IFN-γ (Lin et al., 2016). Furthermore, the engineering of second-generation EE-IFN-γ without site II and III epitopes should eliminate binding to AIGAs, restoring IFN-γ signaling and improving the therapeutic effect in patients (e.g., patients 1 and 3). Alternatively, it is also feasible to engineer autoantigen-based immunoreceptors as chimeric antigen receptors for manipulating T cells to target and deplete autoreactive B lymphocytes in a specific manner (Ellebrecht et al., 2016), rather than using rituximab to target antibody-secreting CD20+ B cells globally (Browne et al., 2012b). The demonstration of Fc-dependent activity for monoclonal AIGAs should therefore drive further studies to investigate the role of this activity in clinical manifestations.
Materials and methods
Subjects
All the participants were adults (>18 yr old), and detailed clinical information for these patients has been reported elsewhere (Chi et al., 2016; Ku et al., 2016). The study was approved by the Institutional Review Board of Chang Gung Memorial Hospital (IRB-102-2095B and IRB-103-2861C), and informed consent was obtained from the patients in accordance with the Declaration of Helsinki.
ELISA
A polystyrene 96-well, flat-bottomed plate (Nunc) was coated with 100 μl IFN-γ (2 μg/ml; R&D Systems) in bicarbonate buffer (pH 9.5) per well and incubated at 4°C overnight. The plate was washed three times in wash buffer (0.05% Tween 20 in PBS) and then blocked by incubation with 5% normal human serum albumin (Aventis) in PBS for 2 h at 25°C. The plate was washed again with wash buffer, and serially diluted plasma samples from healthy donors and patients (dilutions: 1:100, 1:500, and 1:2,500) or mAbs (1 μg/ml) were added to the wells of the plate. The plate was then incubated at 25°C for 2 h, to allow binding to IFN-γ. The plate was washed thoroughly, three times, with wash buffer, and alkaline phosphate-conjugated goat anti-human IgG (1:5,000; Jackson ImmunoResearch Laboratories; for plasma detection)/HRP-conjugated mouse anti-human IgG (1:5,000; Jackson ImmunoResearch Laboratories; for mAb detection) was added. The plate was incubated at 25°C for 1 h and washed five times in wash buffer. For plasma detection, we added p-nitrophenyl phosphate (100 μl/well; Sigma-Aldrich) and incubated the plate at 37°C for 60 min. For mAb detection, we added 3,3′,5,5′-tetramethylbenzidine (TMB) solution (BD Biosciences; 100 μl/well) and incubated the plate at 37°C for 10 min. The reaction was stopped by adding 50 µl 2 N H2SO4. Absorbance was determined at 405 nm for plasma detection or at 450 nm for mAb detection, with a VICTOR X3 Multilabel Plate Reader (PerkinElmer).
Whole-blood activation assay
Blood samples from healthy donors and patients were collected into sterile heparin-coated tubes and mixed with an equal volume of RPMI-1640 (Gibco) supplemented with 1% penicillin/streptomycin. The mixture was dispensed into six wells (1 ml/well) of a 24-well plate and was either left nonactivated (NA) or was activated by incubation with BCG, BCG + IFN-γ (250 ng/ml), or BCG + IL-12 (20 ng/ml; R&D Systems) for 48 h at 37°C under an atmosphere containing 5% CO2. The supernatant was collected from each well and stored at −80°C for cytokine determination.
Cytokine detection in whole blood
Cytokine concentrations were determined by ELISA, with human IL-12p40 kits (BD Biosciences) and human IFN-γ kits (BD Biosciences), according to the manufacturer’s recommendations. Absorbance was determined with a VICTOR X3 Multilabel Plate Reader (PerkinElmer).
Isolation of IFN-γ–specific IgG+ B cells by FACS
Peripheral venous blood samples were collected from human patients with mycobacterial diseases. Mononuclear cells were isolated from these samples by Ficoll-Paque (GE Healthcare) density-gradient centrifugation according to the manufacturer’s instructions. The purified mononuclear cells were collected by centrifugation and resuspended in FACS buffer (1% FBS, 2 mM EDTA, and 0.1% NaN3 in PBS) with 5% normal mouse serum (Jackson ImmunoResearch Laboratories). Background IFN-γR binding was eliminated by blocking the cells by incubation with anti-human CD119 antibody (BioLegend) for 30 min at 4°C. PBMCs (1 × 106 cells) were washed twice with FACS buffer and incubated with 1 μg recombinant human IFN-γ protein for 20 min at 4°C, before staining with anti-human IgG–PE (BD Bioscience), anti-human IgD–APC (BD Bioscience), anti-human CD3–PE-Cyanine7 (eBioscience), anti-human CD19–APC-eFluor 780 (eBioscience), and anti-human IFN-γ–FITC (BD Bioscience) antibodies, and with 7-aminoactinomycin D (Sigma-Aldrich) as a DNA marker. Cells were stained for 30 min at 4°C, and live IgG+IgD−CD19+CD3−IFN-γ+ cells in 96-cell plates were gated and sorted with a FACSAria IIu (Becton Dickinson) for patients 1 and 2 and a MoFlo XDP (Beckman Coulter) for patient 3. All wells contained 18 μl/well lysis buffer with 200 ng random hexamer primer (Thermo Fisher Scientific), 1 μl of a mixture of dNTPs (10 mM each; Thermo Fisher Scientific), 0.5% vol/vol Igepal CA-630 (Sigma-Aldrich), and 40 U Ribolock (Fermentas). A lysate containing total RNA from single B cells was thus obtained. After sorting, plates were sealed with aluminum sealing tape (Corning) and stored at −80°C until further processing.
Antibody heavy/light chain amplification and sequence analysis
The procedure for PCR amplification has been described elsewhere (Chen et al., 2017). Briefly, total RNA from each B cell lysate was subjected to reverse transcription with Maxima H minus reverse transcriptase (Thermo Fisher Scientific) at 42°C for 10 min, 25°C for 10 min, 50°C for 45 min, and 85°C for 5 min. The resulting cDNA was used as a template for amplifying the heavy and light chains with Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific) and V gene-specific primer mixes, in a sequential seminested approach (Chen et al., 2017). PCR products were analyzed by gel electrophoresis to check that the bands obtained were of the correct size, and were then subjected to Sanger sequencing. The sequences obtained were analyzed with IMGT, the international ImMunoGeneTics information system (http://www.imgt.org), to identify the V(D)J gene segments with the highest identity and the numbers of SHMs in VH/VL genes (Lefranc et al., 2009). UCA sequences were determined by reverting mutations to the germline sequence while retaining the original CDR3 junctions and terminal deoxynucleotidyl transferase N nucleotides.
Cloning and expression of purified IgG1 recombinant antibodies
The V(D)J gene fragments were constructed with GeneArt Gene Synthesis (Thermo Fisher Scientific) and inserted into human IgG1 with IgK or IgL expression vectors (Chen et al., 2017). Recombinant antibodies were produced by the transient transfection of Freestyle 293-F cells (Thermo Fisher Scientific) with antibody expression plasmids in the presence of polyethylenimine (Polysciences). After transfection, cells were maintained in FreeStyle 293 Expression Medium (Gibco) at 37°C under an atmosphere containing 8% CO2, with continual shaking at 145 rpm for 3 d. The supernatants were harvested, and the antibodies were isolated on Protein A Sepharose Fast Flow beads (GE Healthcare). The antibodies were eluted from the beads in 0.1 M glycine (pH 2.5) buffered and neutralized with 1 M Tris (pH 8). A buffer exchange to PBS was then performed across a dialysis membrane (Spectra/Por6, 50 kD). Antibody concentrations were determined by UV spectrophotometry (Implen NP80), and antibodies were stored at 4°C until use.
Immunoblotting
Recombinant human IFN-γ protein (300 ng/well) was subjected to SDS-PAGE in a 12% polyacrylamide gel under reducing conditions at 120 V for 2 h. The proteins were then transferred to a polyvinylidene difluoride membrane (Invitrogen Life Technologies) by electroblotting at 250 mA for 2 h. The membrane was washed three times with wash buffer (0.1% Tween 20 in Tris-buffered saline; TBS) and blocked by incubation for 1 h at 4°C with 5% skim milk in wash buffer. The membrane was incubated with 1 μg/ml antibody in blocking solution at 4°C overnight. It was then washed three times with wash buffer and incubated with a 1:5,000 dilution of mouse anti-human IgG conjugated to HRP (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. The membrane was washed three times, and bands were detected by enhanced chemiluminescence (Merck Millipore). Data from a representative experiment are shown.
BLI
Kinetic analysis
Kinetic rate constants (ka and kd) of monoclonal AIGAs to IFN-γ were measured by BLI (FortéBio Octet RED96). The running buffer used contained 0.1% BSA, 0.1% Tween-20, 250 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 in sterile water. Monoclonal AIGAs (1 μg/ml) were loaded onto anti-human IgG Fc capture biosensors (FortéBio) to reach a wavelength shift range of 0.5∼2.0 nm and were then exposed to various concentrations of IFN-γ in running buffer for the association step, before dissociation in running buffer. Rate constants were calculated by fitting a curve (1:1 Langmuir model) of binding response, with the 5-min association and 15-min dissociation interaction times, with Octet Data Analysis software v9.0.
In-tandem cross-competition assay
Biotinylated IFN-γ (biotin–IFN-γ, 2 μg/ml) was loaded onto the streptavidin biosensor (FortéBio) to trigger a saturated response. After 100- and 120-s baseline steps, the primary mAb (5 μg/ml) was individually loaded onto the biosensors in a 600-s association step to achieve increments in wavelength (nm) to saturation. The secondary mAb (5 μg/ml) was incubated with the coated biosensor for 600 s. If a wavelength shift (nm) was observed, it was considered to bind to a different epitope.
Detection of synthetic human IFN-γ peptides by monoclonal AIGAs
The human IFN-γ 30-mer nonoverlapping peptides were synthesized by Kelowna International Scientific, Taiwan. The peptides were lyophilized, and their purity and mass were assessed by HPLC and mass spectrometry, respectively. A transparent 96-well flat-bottomed plate (Nunc) was coated by incubation overnight at 4°C with 100 μl of synthetic IFN-γ peptide (100 μg/ml) in bicarbonate buffer (pH 9.5) per well. The plate was washed three times with wash buffer (0.05% Tween 20 in PBS) and then blocked by incubation with 5% normal human serum albumin (Aventis) in PBS for 2 h at 25°C. The plate was washed again with wash buffer. Each mAb (1 μg/ml) was added to an individual well and allowed to bind to the synthetic IFN-γ peptide for 2 h at 25°C. The plate was washed thoroughly three times in wash buffer, and HRP-conjugated mouse anti-human IgG (1:5,000) was added. The plate was incubated at 25°C for 1 h and then washed five times with wash buffer. We added TMB solution (100 μl/well), and incubated the plate at 37°C for 10 min. The reaction was stopped by adding 50 µl 2N H2SO4. Absorbance was measured at 450 nm with a VICTOR X3 Multilabel Plate Reader (PerkinElmer).
Detection of the recombinant His-tagged IFN-γ variants by monoclonal AIGAs
Recombinant IFN-γ proteins were generated as N-terminally his-tagged constructs in pcDNA3.4 TOPO vector: human IFN-γ (including WT, H19D, and S20P) and human-bovine chimeric proteins (Zuber et al., 2016; bovine C & E: in which the helical regions C and E of human IFN-γ were replaced with the corresponding residues from bovine IFN-γ). Polyethylenimine was used for transfection with the IFN-γ variants, which were expressed in the FreeStyle 293 expression system. The his-tagged proteins from the secreted medium were purified with Ni Sepharose 6 Fast Flow (GE Healthcare), according to the manufacturer’s instructions. A transparent 96-well flat-bottomed plate (Nunc) was coated by incubation overnight at 4°C with 100 μl of IFN-γ variants (1.5 μg/ml) in bicarbonate buffer (pH 9.6) per well. The plate was washed three times with wash buffer (0.05% Tween 20 in PBS) and then blocked by incubation with 1% BSA in PBS for 1 h at 25°C. The plate was washed again with wash buffer. Each mAb (1 μg/ml) was added to an individual well and allowed to bind to the IFN-γ variants for 1 h at 25°C. The plate was washed thoroughly three times in wash buffer, and HRP-conjugated mouse anti-human IgG (1:5,000) was added. The plate was incubated at 37°C for 1 h and then washed five times with wash buffer. We added TMB solution (100 μl/well) and incubated the plate at 37°C for 10 min. The reaction was stopped by adding 50 µl of 2N H2SO4. Absorbance was measured at 450 nm with a VICTOR X3 Multilabel Plate Reader (PerkinElmer).
Particle-based interaction analysis
Bio-plex Pro Magnetic COOH beads (1 × 106 beads/ml; Bio-Rad Laboratories) were coupled with 1 μg IFN-γ, modified as previously described (Ding et al., 2012). Approximately 5,000 IFN-γ–functionalized beads were incubated with the indicated concentration of mAbs (0.01, 0.1, and 1 μg/ml) in PBS. The mixture was shaken at room temperature for 1 h, and the beads were then washed three times in wash buffer (0.05% Tween 20 in PBS). PE-conjugated goat anti-human IgG (eBiosciences) was added (1 μg/ml per well), and the plates were incubated for 40 min at room temperature, with shaking. The beads were washed three times and resuspended in 120 μl PBS; data were acquired with a Bio-Plex 200 instrument (Bio-Rad Laboratories).
In vitro assay of the neutralization of IFN-γ signaling on GAS-luciferase reporter HeLa cells
HeLa cells (BCRC-60005) were transfected with GAS-luciferase reporter plasmids (Promega) and selected on hygromycin B (Sigma-Aldrich). We used 2 × 104 HeLa GAS cells to seed flat 96-well plates. The cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. They were stimulated for 20 h in medium containing IFN-γ (4 ng/ml), with or without threefold serial dilutions of mAbs or control IgG (1 μg/ml). After 20 h of stimulation, luciferase activity was measured with the ONE-Glo luciferase assay system (Promega) according to the manufacturer’s instructions. IFN-γ neutralization was assessed as the percentage decrease in relative luminescence units (RLU), as follows: [1 − (RLU of a single well − RLU of cells grown with 0 ng/ml IFN-γ and 1 μg/ml mAb) × (RLU of cells grown with 4 ng/ml IFN-γ and 0 μg/ml mAb − RLU of cells grown with 0 ng/ml IFN-γ and 1 μg/ml mAb)−1] × 100. The IC50 of each neutralizing mAb was calculated by curve fitting with Prism 9.0 (GraphPad Software).
HLA expression of THP-1 cells
Monoclonal AIGAs or control IgG (3 μg/ml) were incubated with or without IFN-γ (20 ng/ml) for 30 min. We then cultured 1 × 105 THP-1 cells (BCRC-60430) in 96-well culture plates, in 200 µl of RPMI-1640 medium supplemented with 10% FBS, and 1% penicillin/streptomycin for 24 h, with or without stimulation. The cells were washed twice with 1% FBS in PBS. The stimulated and unstimulated THP-1 cells were stained with anti-human HLA-DR–PE antibody (BD Pharmigen) and incubated on ice in the dark for 1 h. The stained cells were washed with 2 ml of 1% FBS in PBS and resuspended in 500 μl of 1% FBS in PBS. The fluorescence intensity of the stained cells was acquired with a FACSVerse flow cytometer (BD Biosciences) and analyzed with FlowJo software v10 (BD Biosciences). The top 5% of fluorescence intensity for unstimulated cells was used as the threshold for positive HLA-DR expression. The inhibitory effect of AIGAs is therefore expressed as the percentage of total THP-1 cells displaying HLA-DR expression relative to the threshold for unstimulated cells.
IFN-γR1 ELISA
A polystyrene 96-well, flat-bottomed plate (Nunc) was coated with 20 nM recombinant human IFN-γR1 protein (R&D Systems) in bicarbonate buffer (pH 9.5) and incubated overnight at 4°C. The plate was washed three times with 0.05% Tween 20 in PBS (wash buffer) and blocked by incubation with 1% BSA in PBS (blocking solution) for 1 h at 25°C. A mixture of mAbs (0, 5, 10, 20, and 100 nM) and biotin–IFN-γ (10 nM) was prepared by incubation for 1 h at 25°C. The plate was washed three times, and the mAb–biotin–IFN-γ mixture was added. The sample was aliquoted into two independent IFN-γR1–coated plates, which were incubated for 1 h at 25°C. The plates were washed three times, and streptavidin-HRP (1:40) was added to one and HRP–conjugated mouse anti-human IgG (1:5,000) in blocking solution to the other. The plates were incubated for 1 h at 25°C and washed three times. We added TMB and incubated the plates at 37°C for 10 min. The reaction was stopped by adding 2N H2SO4. We measured optical density at 450 nm (OD450nm) with a VICTOR X3 Multilabel Plate Reader (PerkinElmer).
Cell-based antibody-binding assay
We incubated 1 × 105 THP-1 cells with 2.5 μg Fc Block (BD Biosciences) in 100 μl RPMI-1640 supplemented with 10% FBS and 1% penicillin/streptomycin for 15 min. We incubated monoclonal AIGAs or control IgG (40 nM) with or without IFN-γ (20 nM) at 25°C for 15 min. The mixtures were then added to the THP-1 cells, which were incubated at 37°C for 15 min. The cells were washed with 2 ml of 1% FBS in PBS, stained by incubation with anti-human IgG-PE (BD Pharmigen) for 30 min at 4°C, and washed with 2 ml of 1% FBS in PBS. The cells were resuspended in 200 μl of 1% FBS in PBS, and the data were collected with a FACSVerse flow cytometer and analyzed with FlowJo software v10.
On-cell IFN-γ receptor dimerization
Two-color single-molecule cotracking experiments were used to quantify receptor homo- and heterodimerization, as previously described (Mendoza et al., 2019). For heterodimerization analyses, HeLa cells were transiently transfected with IFN-γR1 and IFN-γR2 N-terminally fused to engineered, nonfluorescent variants of monomeric GFP, mXFPe, and mXFPm, respectively. Photostable dyes were used for selective labeling, with the anti-GFP nanobodies Enhancer and Minimizer (Kirchhofer et al., 2010) conjugated to ATTO 643 and Rho11, respectively, via a single cysteine residue. For homodimerization analyses, HeLa cells were transiently transfected with mXFPe-IFN-γR2 and labeled with anti-GFP nanobodies Minimizer conjugated to ATTO 643 and Rho 11, in equal proportions. We incubated 20 nM mAb and 10 nM IFN-γ on ice for 30 min, before adding this mixture to the culture medium. For ligand-binding experiments, 5 nM IFN-γ labeled on S66C with either Rho 11 or DY 647 was incubated with 20 nM mAb for 30 min on ice and applied to untransfected HeLa cells. Time-lapse dual-color imaging was performed by TIRF microscopy, with excitation at 561 and 640 nm and detection with a single EMCCD camera using a spectral image splitter. Molecules were localized with a multiple-target tracing algorithm (Serge et al., 2008). Receptor dimers were identified as molecules colocalizing within 100 nm of each other for ≥10 consecutive frames (Wilmes et al., 2015).
Binding and dimerization of IFN-γ at the cell surface
For the site-specific labeling of IFN-γ, an additional cysteine residue was introduced by mutation of the exposed serine residue, S66. IFN-γ S66C was produced in Escherichia coli strain BL21 RIL and purified to homogeneity from inclusion bodies, as previously described (Nuara et al., 2008). Purified IFN-γ S66C was labeled in a site-specific manner with ATTO Rho11 (ATTO-TEC) or DY647P1 (Dyomics) maleimide conjugates. To this end, 500 µl of 35 µM protein was incubated with 105 µM dye at room temperature for 45 min. The reaction was stopped by incubation with 300 µM L-cysteine for 15 min. Finally, free dye was removed by SEC (Superdex 75 10/300; GE Healthcare). Under these coupling conditions, ∼95% labeling was achieved, as confirmed by UV/Vis spectroscopy. For the quantification of IFN-γ binding and dimerization on the cell surface, WT HeLa cells were incubated with IFN-γ Rho11 and IFN-γ DY647P1, each at a concentration of 5 nM, for 5 min at room temperature. Single-molecule imaging and analysis were performed as described in the main methods section.
SEC
Equimolar mixtures of AIGAs or control IgG (3 μM) and IFN-γ (3 μM) were prepared in PBS and incubated for 1 h at room temperature. Samples (40 μl) were injected and analyzed on a Waters Acquity Arc Bio UHPLC machine with an XBridge Protein BEH SEC column (Waters Corp.; 450 Å, 2.5 μm, 4.6 mm × 150 mm) and an Arg-SEC mobile phase (COSMOSIL; flow rate: 0.3 ml/min). BEH450 SEC Protein Standard Mix (Waters Corp.) was injected to monitor protein separation. Detection was performed with a PDA detector (Waters Corp.) with absorption at 280 nm.
Luciferase assay of Fc receptor activation
Jurkat cells (BCRC-60424) were stably transfected with the FcγIIIa V158 variant and nuclear factor of activated T cells response element (NFAT-RE) luciferase constructs and selected on geneticin and hygromycin B, for use as effector cells. The biological activity of antibodies in ADCC was quantified by assessing luciferin production following NFAT pathway activation. The human monocyte cell line THP-1 expressing IFN-γR was used as the target cells. We seeded 96-well plates with effector and target cells in a ratio of 5:1 (total 6 × 104 cells). The cells were incubated with mAbs, LALA variants, or control IgG (10 nM) and IFN-γ (0, 5 and 10 nM) for 6 h at 37°C. Luciferase activity, indicating Fc receptor activation, was then assessed with the ONE-Glo luciferase assay system (Promega) according to the manufacturer’s instructions.
Cytotoxicity assay
Monocytes and NK cells were isolated from whole blood from healthy donors with RosetteSep (STEMCELL Technologies), in accordance with the manufacturer’s instructions. Isolated NK cells (effector cells) and monocytes (target cells) were mixed in a 10:1 ratio (total 5.5 × 104 cells) and cultured with the indicated concentration of mAbs or LALA variants (10 nM), with or without IFN-γ (10 nM). The cells were incubated at 37°C for 8 h, and cytotoxicity was then assessed with the CytoTox-Glo Cytotoxicity Assay (Promega), according to the manufacturer’s instructions. Cellular cytotoxicity was calculated as the percentage increase in RLU, as follows: [RLU of single well – RLU of cells (E:T 10:1) incubated with 10 nM IFN-γ and 0 nM of the corresponding mAb] × [RLU of total lysed cells (E:T 0:1) − RLU of cells for (E:T 0:1)]−1.
IL-12p40 and CXCL-10 production
PBMCs from healthy donors were cultured at a density of 1 × 106 cells/ml in RPMI-1640 supplemented with 10% FBS and 1% penicillin/streptomycin. They were either left nonactivated or were treated with BCG, BCG + IFN-γ (25 ng/ml) in the presence of AIGAs, or control IgG (1 μg/ml) previously incubated at room temperature for 30 min. After incubation for 48 h, the supernatants were collected and subjected to ELISA for human IL-12p40 (BD Biosciences) and human CXCL-10 (R&D Systems) according to the kit manufacturers’ instructions.
Statistical analysis
All statistical analyses were performed with Prism software (GraphPad v9). The sample size (n) for experiments and the statistical tests used are indicated in the corresponding figure legends. P values ≤0.05 were considered significant: *, P < 0.05; **, P < 0.01; ***, P < 0.005; and ****, P < 0.001.
Online supplemental material
Fig. S1 shows the generation of fully human pathogenic monoclonal AIGAs isolated from three recruited patients with mycobacterial diseases. Fig. S2 shows the kinetic response of antibody to IFN-γ and the identification of epitopes for monoclonal AIGAs against IFN-γ. Fig. S3 indicates the neutralization of IFN-γ in vitro for different capacities of monoclonal AIGAs. Fig. S4 demonstrates the cross-linking of surface-bound IFN-γ by monoclonal AIGAs. Fig. S5 summarizes the properties of 19 monoclonal AIGAs and AMG811 based on their binding sites on IFN-γ.
Supplementary Material
contains original blots for Fig. S1.
Acknowledgments
We thank the patients’ families for their trust and cooperation. We thank S.-C. Jao and W.-C. Huang (Academia Sinica), Z.-Q. Gu and Y.-S. Chu (National Yang Ming Chiao Tung University), and C.-L. Hsiao and Y.-T. Kuo (Elixiron Immunotherapeutics) for providing advice concerning experiments. We thank the Flow Cytometric Analyzing and Sorting Core Facility at National Taiwan University Hospital. We thank Mark R. Walter (University of Alabama at Birmingham, Birmingham, AL) for providing a plasmid for producing IFN-γ.
The study was funded by Chang Gung Memorial Hospital (CMRPD1K0681-2 and BMRPB98), the Taiwan Ministry of Science and Technology (105-2628-B-182-002-MY3) and the Deutsche Forschungsgemeinschaft (PI 405/14-1 and INST 190/146-3). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Author contributions: C.-L. Ku conceptualized the study; H.-P. Shih, J.-Y. Ding, and C.-H. Lin conceived the experiments with input from C.-L. Ku, who supervised the project. H.-P. Shih, J.-Y. Ding, and C.-H. Lin planned the experiments and analyzed the results. Y.-M. Liu, T.-S. Wu, and C.-Y. Chi provided the patient samples. H.-P. Shih carried out the bulk of the experiments. H.-T. Ting, T.-Y. Wu, and Y.-N. Lin prepared and purified the monoclonal antibodies. J. Sotolongo Bellón performed and analyzed the single-molecule tracking experiment, supervised by J. Piehler. H.-P. Shih performed the cytotoxicity assays with the help of Y.-F. Lo and analyzed the results of these experiments with Y.-H. Tsai. H.-P. Shih and C.-L. Ku wrote the manuscript. All the authors discussed the results, provided critical feedback, contributed to the writing of the manuscript, or revised and commented on the manuscript.
References
- Aoki, A., Sakagami T., Yoshizawa K., Shima K., Toyama M., Tanabe Y., Moro H., Aoki N., Watanabe S., Koya T., et al. 2018. Clinical significance of interferon-γ neutralizing autoantibodies against disseminated nontuberculous mycobacterial disease. Clin. Infect. Dis. 66:1239–1245. 10.1093/cid/cix996 [DOI] [PubMed] [Google Scholar]
- Bach, E.A., Szabo S.J., Dighe A.S., Ashkenazi A., Aguet M., Murphy K.M., and Schreiber R.D.. 1995. Ligand-induced autoregulation of IFN-γ receptor β chain expression in T helper cell subsets. Science. 270:1215–1218. 10.1126/science.270.5239.1215 [DOI] [PubMed] [Google Scholar]
- Bastard, P., Orlova E., Sozaeva L., Levy R., James A., Schmitt M.M., Ochoa S., Kareva M., Rodina Y., Gervais A., et al. 2021. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 218:e20210554. 10.1084/jem.20210554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard, P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V., et al. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370:eabd4585. 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedigheimer, M.J., Martin D.A., Amoura Z., Sanchez-Guerrero J., Romero-Diaz J., Kivitz A., Aranow C., Chan T.M., Chong Y.B., Chiu K., et al. 2017. Safety, pharmacokinetics and pharmacodynamics of AMG 811, an anti-interferon-γ monoclonal antibody, in SLE subjects without or with lupus nephritis. Lupus Sci. Med. 4:e000226. 10.1136/lupus-2017-000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dupuis, S., and Bustamante J.. 2021. Mycobacterial diseases in patients with inborn errors of immunity. Curr. Opin. Immunol. 72:262–271. 10.1016/j.coi.2021.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne, S.K., Burbelo P.D., Chetchotisakd P., Suputtamongkol Y., Kiertiburanakul S., Shaw P.A., Kirk J.L., Jutivorakool K., Zaman R., Ding L., et al. 2012a. Adult-onset immunodeficiency in Thailand and Taiwan. N. Engl. J. Med. 367:725–734. 10.1056/NEJMoa1111160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne, S.K., Zaman R., Sampaio E.P., Jutivorakool K., Rosen L.B., Ding L., Pancholi M.J., Yang L.M., Priel D.L., Uzel G., et al. 2012b. Anti-CD20 (rituximab) therapy for anti-IFN-γ autoantibody-associated nontuberculous mycobacterial infection. Blood. 119:3933–3939. 10.1182/blood-2011-12-395707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, J. 2020. Mendelian susceptibility to mycobacterial disease: Recent discoveries. Hum. Genet. 139:993–1000. 10.1007/s00439-020-02120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, J., Boisson-Dupuis S., Abel L., and Casanova J.L.. 2014. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol. 26(6):454–470. . 10.1016/j.smim.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J.F., Trendell-Smith N.J., Chan J.C., Hung I.F., Tang B.S., Cheng V.C., Yeung C.K., and Yuen K.Y.. 2013. Reactive and infective dermatoses associated with adult-onset immunodeficiency due to anti-interferon-gamma autoantibody: Sweet’s syndrome and beyond. Dermatology. 226:157–166. 10.1159/000347112 [DOI] [PubMed] [Google Scholar]
- Chaowattanapanit, S., Choonhakarn C., Chetchotisakd P., Sawanyawisuth K., and Julanon N.. 2016. Clinical features and outcomes of Sweet’s syndrome associated with non-tuberculous mycobacterial infection and other associated diseases. J. Dermatol. 43:532–536. 10.1111/1346-8138.13167 [DOI] [PubMed] [Google Scholar]
- Chen, F., Tzarum N., Wilson I.A., and Law M.. 2019. VH1-69 antiviral broadly neutralizing antibodies: Genetics, structures, and relevance to rational vaccine design. Curr. Opin. Virol. 34:149–159. 10.1016/j.coviro.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.B., James L.K., Davies A.M., Wu Y.B., Rimmer J., Lund V.J., Chen J.H., McDonnell J.M., Chan Y.C., Hutchins G.H., et al. 2017. Antibodies and superantibodies in patients with chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 139:1195–1204.e11. 10.1016/j.jaci.2016.06.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, C.Y., Chu C.C., Liu J.P., Lin C.H., Ho M.W., Lo W.J., Lin P.C., Chen H.J., Chou C.H., Feng J.Y., et al. 2013. Anti-IFN-γ autoantibodies in adults with disseminated nontuberculous mycobacterial infections are associated with HLA-DRB1*16:02 and HLA-DQB1*05:02 and the reactivation of latent varicella-zoster virus infection. Blood. 121:1357–1366. 10.1182/blood-2012-08-452482 [DOI] [PubMed] [Google Scholar]
- Chi, C.Y., Lin C.H., Ho M.W., Ding J.Y., Huang W.C., Shih H.P., Yeh C.F., Fung C.P., Sun H.Y., Huang C.T., et al. 2016. Clinical manifestations, course, and outcome of patients with neutralizing anti-interferon-γ autoantibodies and disseminated nontuberculous mycobacterial infections. Medicine. 95:e3927. 10.1097/MD.0000000000003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes, R., and Ravetch J.V.. 1995. Cytotoxic antibodies trigger inflammation through Fc receptors. Immunity. 3:21–26. 10.1016/1074-7613(95)90155-8 [DOI] [PubMed] [Google Scholar]
- Ding, L., Mo A., Jutivorakool K., Pancholi M., Holland S.M., and Browne S.K.. 2012. Determination of human anticytokine autoantibody profiles using a particle-based approach. J. Clin. Immunol. 32:238–245. 10.1007/s10875-011-9621-8 [DOI] [PubMed] [Google Scholar]
- Doffinger, R., Helbert M.R., Barcenas-Morales G., Yang K., Dupuis S., Ceron-Gutierrez L., Espitia-Pinzon C., Barnes N., Bothamley G., Casanova J.L., et al. 2004. Autoantibodies to interferon-gamma in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin. Infect. Dis. 38:e10–e14. 10.1086/380453 [DOI] [PubMed] [Google Scholar]
- Dupuis, S., Doffinger R., Picard C., Fieschi C., Altare F., Jouanguy E., Abel L., and Casanova J.L.. 2000. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol. Rev. 178:129–137. 10.1034/j.1600-065x.2000.17810.x [DOI] [PubMed] [Google Scholar]
- Ellebrecht, C.T., Bhoj V.G., Nace A., Choi E.J., Mao X., Cho M.J., Di Zenzo G., Lanzavecchia A., Seykora J.T., Cotsarelis G., et al. 2016. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science. 353:179–184. 10.1126/science.aaf6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, E., Gilman M.S.A., Wrapp D., Chen M., Ngwuta J.O., Moin S.M., Bai P., Sivasubramanian A., Connor R.I., Wright P.F., et al. 2018. Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation. Immunity. 48:339–349.e5. 10.1016/j.immuni.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlund, A.C., Schreiber R.D., Goeddel D.V., and Pennica D.. 1993. Interferon-gamma induces receptor dimerization in solution and on cells. J. Biol. Chem. 268:18103–18110. 10.1016/s0021-9258(17)46817-7 [DOI] [PubMed] [Google Scholar]
- Guo, J., Ning X.Q., Ding J.Y., Zheng Y.Q., Shi N.N., Wu F.Y., Lin Y.K., Shih H.P., Ting H.T., Liang G., et al. 2020. Anti-IFN-γ autoantibodies underlie disseminated Talaromyces marneffei infections. J. Exp. Med. 217:e20190502. 10.1084/jem.20190502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell, A.J., Hangartner L., Hunter M., Havenith C.E., Beurskens F.J., Bakker J.M., Lanigan C.M., Landucci G., Forthal D.N., Parren P.W., et al. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 449:101–104. 10.1038/nature06106 [DOI] [PubMed] [Google Scholar]
- Hezareh, M., Hessell A.J., Jensen R.C., van de Winkel J.G., and Parren P.W.. 2001. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J. Virol. 75:12161–12168. 10.1128/JVI.75.24.12161-12168.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoflich, C., Sabat R., Rosseau S., Temmesfeld B., Slevogt H., Docke W.D., Grutz G., Meisel C., Halle E., Gobel U.B., et al. 2004. Naturally occurring anti-IFN-γ autoantibody and severe infections with Mycobacterium cheloneae and Burkholderia cocovenenans. Blood. 103:673–675. 10.1182/blood-2003-04-1065 [DOI] [PubMed] [Google Scholar]
- Hong, G.H., Ortega-Villa A.M., Hunsberger S., Chetchotisakd P., Anunnatsiri S., Mootsikapun P., Rosen L.B., Zerbe C.S., and Holland S.M.. 2020. Natural history and evolution of anti-interferon-γ autoantibody-associated immunodeficiency syndrome in Thailand and the United States. Clin. Infect. Dis. 71:53–62. 10.1093/cid/ciz786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X., and Ivashkiv L.B.. 2009. Cross-regulation of signaling pathways by interferon-gamma: Implications for immune responses and autoimmune diseases. Immunity. 31:539–550. 10.1016/j.immuni.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv, L.B. 2018. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 18:545–558. 10.1038/s41577-018-0029-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis, R., Lund J., and Pound J.. 1990. Molecular definition of interaction sites on human IgG for Fc receptors (huFc gamma R). Mol. Immunol. 27:1237–1240. 10.1016/0161-5890(90)90027-w [DOI] [PubMed] [Google Scholar]
- Jutivorakool, K., Sittiwattanawong P., Kantikosum K., Hurst C.P., Kumtornrut C., Asawanonda P., Klaewsongkram J., and Rerknimitr P.. 2018. Skin manifestations in patients with adult-onset immunodeficiency due to anti-interferon-gamma autoantibody: A relationship with systemic infections. Acta Derm. Venereol. 98:742–747. 10.2340/00015555-2959 [DOI] [PubMed] [Google Scholar]
- Kampitak, T., Suwanpimolkul G., Browne S., and Suankratay C.. 2011. Anti-interferon-γ autoantibody and opportunistic infections: Case series and review of the literature. Infection. 39:65–71. 10.1007/s15010-010-0067-3 [DOI] [PubMed] [Google Scholar]
- Kampmann, B., Hemingway C., Stephens A., Davidson R., Goodsall A., Anderson S., Nicol M., Scholvinck E., Relman D., Waddell S., et al. 2005. Acquired predisposition to mycobacterial disease due to autoantibodies to IFN-gamma. J. Clin. Invest. 115:2480–2488. 10.1172/JCI19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer, A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C.S., Cardoso M.C., et al. 2010. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17:133–138. 10.1038/nsmb.1727 [DOI] [PubMed] [Google Scholar]
- Kreer, C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. 2020. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 182:1663–1673. 10.1016/j.cell.2020.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, C.L., Chi C.Y., von Bernuth H., and Doffinger R.. 2020. Autoantibodies against cytokines: Phenocopies of primary immunodeficiencies?. Hum. Genet. 139:783–794. 10.1007/s00439-020-02180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, C.L., Lin C.H., Chang S.W., Chu C.C., Chan J.F., Kong X.F., Lee C.H., Rosen E.A., Ding J.Y., Lee W.I., et al. 2016. Anti-IFN-γ autoantibodies are strongly associated with HLA-DR*15:02/16:02 and HLA-DQ*05:01/05:02 across Southeast Asia. J. Allergy Clin. Immunol. 137:945–948.e8. 10.1016/j.jaci.2015.09.018 [DOI] [PubMed] [Google Scholar]
- Lefranc, M.P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. 2009. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 37:D1006–D1012. 10.1093/nar/gkn838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.H., Chi C.Y., Shih H.P., Ding J.Y., Lo C.C., Wang S.Y., Kuo C.Y., Yeh C.F., Tu K.H., Liu S.H., et al. 2016. Identification of a major epitope by anti-interferon-γ autoantibodies in patients with mycobacterial disease. Nat. Med. 22:994–1001. 10.1038/nm.4158 [DOI] [PubMed] [Google Scholar]
- Lundell, D.L., and Narula S.K.. 1994. Structural elements required for receptor recognition of human interferon-gamma. Pharmacol. Ther. 64:1–21. 10.1016/0163-7258(94)90031-0 [DOI] [PubMed] [Google Scholar]
- Luster, A.D., and Ravetch J.V.. 1987. Biochemical characterization of a gamma interferon-inducible cytokine (IP-10). J. Exp. Med. 166:1084–1097. 10.1084/jem.166.4.1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan, C., Fieschi C., Lammas D.A., Picard C., Dorman S.E., Sanal O., MacLennan J.M., Holland S.M., Ottenhoff T.H., Casanova J.L., and Kumararatne D.S.. 2004. Interleukin (IL)-12 and IL-23 are key cytokines for immunity against Salmonella in humans. J. Infect. Dis. 190:1755–1757. 10.1086/425021 [DOI] [PubMed] [Google Scholar]
- Meffre, E., and Wardemann H.. 2008. B-cell tolerance checkpoints in health and autoimmunity. Curr. Opin. Immunol. 20:632–638. 10.1016/j.coi.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Mendoza, J.L., Escalante N.K., Jude K.M., Sotolongo Bellon J., Su L., Horton T.M., Tsutsumi N., Berardinelli S.J., Haltiwanger R.S., Piehler J., et al. 2019. Structure of the IFNγ receptor complex guides design of biased agonists. Nature. 567:56–60. 10.1038/s41586-019-0988-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn, F., and Ravetch J.V.. 2005. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 310:1510–1512. 10.1126/science.1118948 [DOI] [PubMed] [Google Scholar]
- Noma, K., Mizoguchi Y., Tsumura M., and Okada S.. 2022. Mendelian susceptibility to mycobacterial diseases: State of the art. Clin. Microbiol. Infect. S1198-743X(22)00143-4. 10.1016/j.cmi.2022.03.004 [DOI] [PubMed] [Google Scholar]
- Nuara, A.A., Walter L.J., Logsdon N.J., Yoon S.I., Jones B.C., Schriewer J.M., Buller R.M., and Walter M.R.. 2008. Structure and mechanism of IFN-γ antagonism by an orthopoxvirus IFN-γ-binding protein. Proc. Natl. Acad. Sci. USA. 105:1861–1866. 10.1073/pnas.0705753105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell, M.A., Luo Y., Chen X., Szilvasi A., Hunter S.E., and Clinton S.K.. 1999. Role of IL-12 in the induction and potentiation of IFN-γ in response to bacillus Calmette-Guérin. J. Immunol. 163:4246–4252 [PubMed] [Google Scholar]
- Parekh, B.S., Berger E., Sibley S., Cahya S., Xiao L., LaCerte M.A., Vaillancourt P., Wooden S., and Gately D.. 2012. Development and validation of an antibody-dependent cell-mediated cytotoxicity-reporter gene assay. MAbs. 4:310–318. 10.4161/mabs.19873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S.Y., Ding L., Brown M.R., Lantz L., Gay T., Cohen S., Martyak L.A., Kubak B., and Holland S.M.. 2005. Anti-IFN-γ autoantibodies in disseminated nontuberculous mycobacterial infections. J. Immunol. 175:4769–4776. 10.4049/jimmunol.175.7.4769 [DOI] [PubMed] [Google Scholar]
- Piccoli, L., Campo I., Fregni C.S., Rodriguez B.M., Minola A., Sallusto F., Luisetti M., Corti D., and Lanzavecchia A.. 2015. Neutralization and clearance of GM-CSF by autoantibodies in pulmonary alveolar proteinosis. Nat. Commun. 6:7375. 10.1038/ncomms8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel, A., Doffinger R., Natividad A., Chrabieh M., Barcenas-Morales G., Picard C., Cobat A., Ouachee-Chardin M., Toulon A., Bustamante J., et al. 2010. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J. Exp. Med. 207:291–297. 10.1084/jem.20091983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel, A., Picard C., Lorrot M., Pons C., Chrabieh M., Lorenzo L., Mamani-Matsuda M., Jouanguy E., Gendrel D., and Casanova J.L.. 2008. Recurrent staphylococcal cellulitis and subcutaneous abscesses in a child with autoantibodies against IL-6. J. Immunol. 180:647–654. 10.4049/jimmunol.180.1.647 [DOI] [PubMed] [Google Scholar]
- Renner, M., Flanagan A., Dejnirattisai W., Puttikhunt C., Kasinrerk W., Supasa P., Wongwiwat W., Chawansuntati K., Duangchinda T., Cowper A., et al. 2018. Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus. Nat. Immunol. 19:1248–1256. 10.1038/s41590-018-0227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveille, J.D. 2006. The genetic basis of autoantibody production. Autoimmun. Rev. 5:389–398. 10.1016/j.autrev.2005.10.012 [DOI] [PubMed] [Google Scholar]
- Rosain, J., Kong X.F., Martinez-Barricarte R., Oleaga-Quintas C., Ramirez-Alejo N., Markle J., Okada S., Boisson-Dupuis S., Casanova J.L., and Bustamante J.. 2019. Mendelian susceptibility to mycobacterial disease: 2014-2018 update. Immunol. Cell Biol. 97:360–367. 10.1111/imcb.12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, K., Hertzog P.J., Ravasi T., and Hume D.A.. 2004. Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 75:163–189. 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- Seelig, G.F., Wijdenes J., Nagabhushan T.L., and Trotta P.P.. 1988. Evidence for a polypeptide segment at the carboxyl terminus of recombinant human gamma interferon involved in expression of biological activity. Biochemistry. 27:1981–1987. 10.1021/bi00406a026 [DOI] [PubMed] [Google Scholar]
- Serge, A., Bertaux N., Rigneault H., and Marguet D.. 2008. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 5:687–694. 10.1038/nmeth.1233 [DOI] [PubMed] [Google Scholar]
- Shih, H.P., Ding J.Y., Yeh C.F., Chi C.Y., and Ku C.L.. 2021. Anti-interferon-γ autoantibody-associated immunodeficiency. Curr. Opin. Immunol. 72:206–214. 10.1016/j.coi.2021.05.007 [DOI] [PubMed] [Google Scholar]
- Tham, E.H., Huang C.H., Soh J.Y., Thayalasingam M., Lee A.J., Lum L.H., Poon L.M., Lye D.C., Chai L.Y., Tambyah P.A., et al. 2016. Neutralizing anti-interferon-gamma autoantibody levels may not correlate with clinical course of disease. Clin. Infect. Dis. 63:572–573. 10.1093/cid/ciw351 [DOI] [PubMed] [Google Scholar]
- Tiller, T., Tsuiji M., Yurasov S., Velinzon K., Nussenzweig M.C., and Wardemann H.. 2007. Autoreactivity in human IgG+ memory B cells. Immunity. 26:205–213. 10.1016/j.immuni.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, K., Nakata K., Trapnell B.C., Terakawa T., Hamano E., Mikami A., Matsushita I., Seymour J.F., Oh-Eda M., Ishige I., et al. 2004. High-affinity autoantibodies specifically eliminate granulocyte-macrophage colony-stimulating factor activity in the lungs of patients with idiopathic pulmonary alveolar proteinosis. Blood. 103:1089–1098. 10.1182/blood-2003-05-1565 [DOI] [PubMed] [Google Scholar]
- Vogt, G., Chapgier A., Yang K., Chuzhanova N., Feinberg J., Fieschi C., Boisson-Dupuis S., Alcais A., Filipe-Santos O., Bustamante J., et al. 2005. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 37:692–700. 10.1038/ng1581 [DOI] [PubMed] [Google Scholar]
- Walter, M.R., Windsor W.T., Nagabhushan T.L., Lundell D.J., Lunn C.A., Zauodny P.J., and Narula S.K.. 1995. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature. 376:230–235. 10.1038/376230a0 [DOI] [PubMed] [Google Scholar]
- Wilkinson, I., Anderson S., Fry J., Julien L.A., Neville D., Qureshi O., Watts G., and Hale G.. 2021. Fc-engineered antibodies with immune effector functions completely abolished. PLoS One. 16:e0260954. 10.1371/journal.pone.0260954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes, S., Beutel O., Li Z., Francois-Newton V., Richter C.P., Janning D., Kroll C., Hanhart P., Hotte K., You C., et al. 2015. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J. Cell Biol. 209:579–593. 10.1083/jcb.201412049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipasa, J., Chaiwarith R., Chawansuntati K., Praparattanapan J., Rattanathammethee K., and Supparatpinyo K.. 2018. Characterization of anti-interferon-γ antibodies in HIV-negative immunodeficient patients infected with unusual intracellular microorganisms. Exp. Biol. Med. 243:621–626. 10.1177/1535370218764086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber, B., Rudstrom K., Ehrnfelt C., and Ahlborg N.. 2016. Epitope mapping of neutralizing monoclonal antibodies to human interferon-γ using human-bovine interferon-γ chimeras. J. Interferon Cytokine Res. 36:542–551. 10.1089/jir.2016.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. S1.