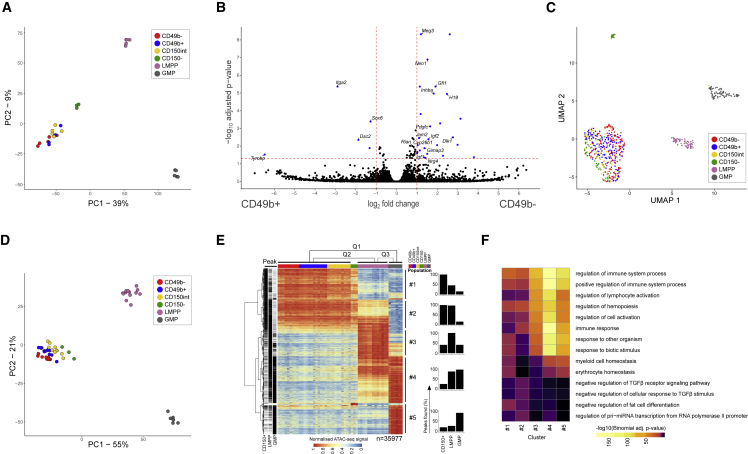
Figure 6.
The CD49b− and CD49b+ subsets are transcriptionally similar but epigenetically distinct
(A) PCA of bulk RNA-seq data (nCD49b− = 5, nCD49b+ = 4, nCD150int = 5, nCD150− = 3, nLMPP = 5, nGMP = 5 replicates, five independent experiments).
(B) Volcano plot of differential expression between CD49b− and CD49b+ subsets. Differentially expressed genes (padj < 0.05, |fold change| > 2) are marked in blue, with selected genes annotated.
(C) UMAP visualization of single-cell RNA-seq data colored by cell population (nCD49b− = 135, nCD49b+ = 146, nCD150int = 59, nCD150− = 77, nLMPP = 57, nGMP = 74 cells, two independent experiments).
(D) PCA of ATAC-seq data (nCD49b− = 9, nCD49b+ = 12, nCD150int = 10, nCD150− = 3, nLMPP = 13, nGMP = 6 replicates, seven independent experiments).
(E) Heatmap (left) of row-normalized chromatin accessibility for differential regions (padj < 1 × 10−4, |fold change| > 2) in pairwise comparisons between CD150+, LMPP, and GMP populations. Regions are divided into five clusters based on hierarchical clustering. Bar graphs (right) show how many peaks were called in each cluster.
(F) GO enrichment analysis of clusters 1–5. The top three significantly enriched terms for each cluster from the GO biological process are shown. See also Figures S6 and S7.