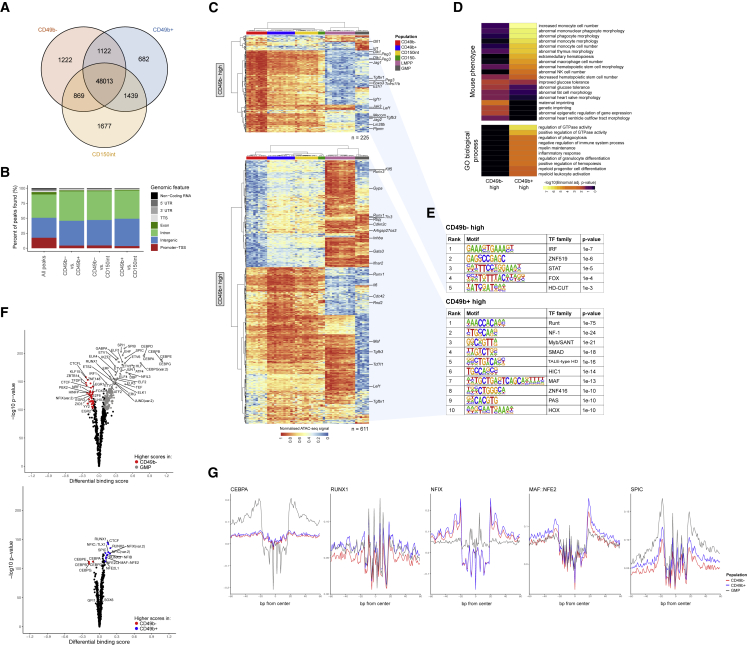
Figure 7.
The CD49b− and CD49b+ subsets exhibit distinct epigenetic changes
(A) Overlap of open chromatin regions between CD150+ populations. Peaks with a normalized read count of >1.5 in more than one-third of samples were considered found.
(B) Genomic feature distribution of regions with differential accessibility (padj < 0.01) and all regions as a reference ("all peaks").
(C) Heatmap of row-normalized chromatin accessibility for regions with differential accessibility (padj < 0.01) between CD49b− (top; n = 225 peaks) and CD49b+ (bottom; n = 611 peaks) subsets. For selected regions the nearest gene is indicated.
(D) GO enrichment analysis of regions with differential accessibility between CD49b− and CD49b+ subsets. The top 10 significantly enriched terms from the mouse phenotype and GO biological process are shown.
(E) Transcription factor (TF) families with enriched binding motifs (q <0.01, top 10) in regions with increased accessibility in CD49b− (top) or CD49b+ (bottom) cells.
(F) Volcano plots of differential TF binding. Transcription factors with differential binding activity (differential binding score >0.1, p < 1 × 10−100) are colored and selectively annotated.
(G) Aggregated footprint plots for TFs with differential binding. See also Figure S7.