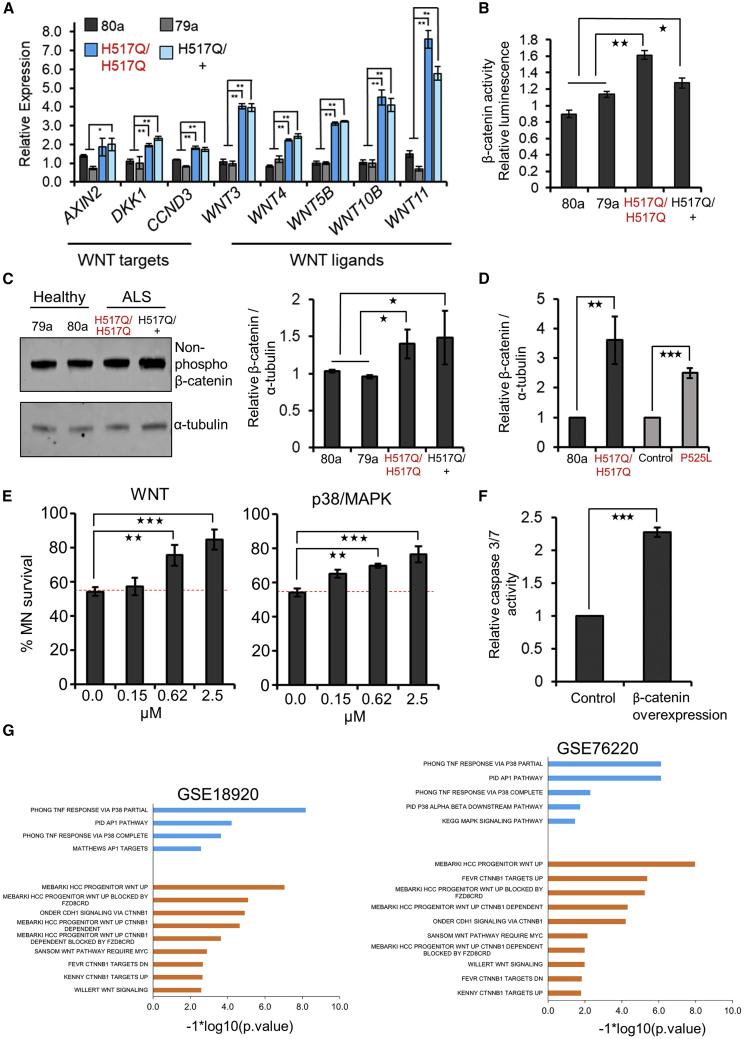
Figure 4.
ALS MNs display increased WNT
(A) qRT-PCR analysis of mRNA expression of WNT genes and ligands in FUS ALS MNs compared to healthy controls (79a and 80a). N = 3.
(B) SuperTopFlash luciferase assay showing relative luciferase units for β-catenin activity in H517Q/H517Q and H517Q/+ MN relative to 80a and 79a healthy controls. N = 3–4.
(C) Representative western blot (left panel) and quantification (right panel) of active β-catenin protein in healthy and FUS ALS MNs. β-catenin normalized to α-tubulin and shown relative to healthy controls. N = 3.
(D) In-cell protein quantification of β-catenin in FUS H517Q/H517Q MN relative to healthy control 80a (N = 5) and FUS P525L MN relative to isogenic control (N = 3).
(E) Quantification of ISL1+ FUS H517Q/H517Q MNs at day 45 relative to day 30 after treatment with small-molecule inhibitors of the indicated pathways. Horizontal dotted line indicates MN loss in the DMSO-only control. N = 3, p values were estimated using Student’s 2-tailed t test.
(F) Caspase-3/7 activity relative to viable cells in response to overexpression of β-catenin versus GFP control in day 23 healthy (80A) MNs. N = 3.
(G) GSEA on sALS MN datasets using gene sets relevant to WNT or p38/MAPK signaling. Blue bars indicate p38/MAPK gene sets. Orange bars indicate WNT/CTNNB1 gene sets. p values were estimated using 1,000 random simulations and adjusted using the Benjamini-Hochberg procedure. N: number of independent differentiations. Error bars indicate SEMs. p values were estimated by Student’s 1-tailed t test unless otherwise indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.