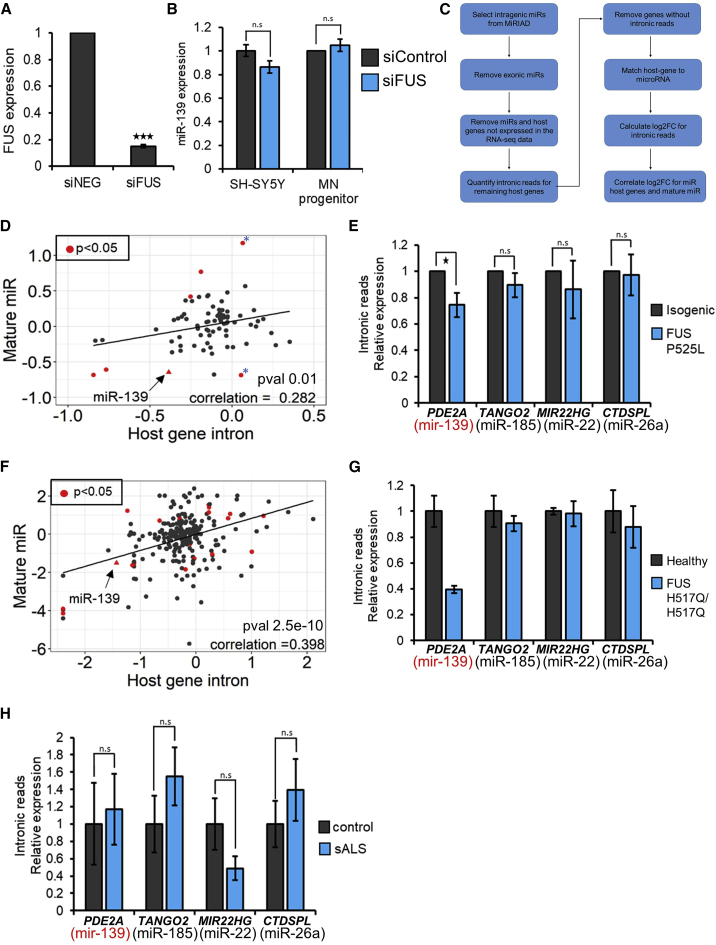
Figure 5.
Mutant FUS transcriptionally downregulates miR-139 potentially through a gain-of-function mechanism
(A) Relative FUS gene expression after siRNA-mediated knockdown in SH-SY5Y cells. Fold changes were normalized to scrambled (negative control) siRNA. N = 3 independent transfections. p values were estimated using Student’s 1-tailed t test.
(B) qRT-PCR analysis of miR-139-5p expression in response to siRNA-mediated FUS knockdown in SH-SY5Y cells and MN progenitors derived from healthy (80a) iPSC. Gray bars: scrambled siRNA used as a negative control; blue bars: siRNA against FUS. N = 3. Fold changes were normalized to scrambled siRNA.
(C) Flowchart depicting the correlation analysis between expression changes in intronic miRs and their host genes.
(D) Scatterplot contrasting log2 fold changes in intronic reads of a host gene versus the corresponding mature miR in FUS P525L MNs relative to the isogenic control MNs. Number of miRs = 80, N = 3. Shown in red are miRNAs identified as differentially expressed in FUS P525L MN at day 19 (p < 0.05, data from our previous publication; De Santis et al., 2017). Blue asterisks indicate example miRNAs described in Mutant FUS downregulates miR-139 potentially by a gain-of-function mechanism.
(E) Relative expression of pri-miRNAs for miR-139-5p, miR-185, miR-22 and miR-26a in FUS P525L MNs. Intronic counts for the isogenic sample were set to 1. N = 3.
(F) Scatterplot contrasting log2 fold changes in intronic reads of host genes versus the corresponding mature miRs in FUS H517Q MNs relative to the healthy control MNs. Number of miRs = 208, N = 2. Shown in red are miRNAs identified as differentially expressed in FUS H517Q MN at day 30 (adjusted p < 0.05).
(G) Relative expression of pri-miRNAs for miR-139-5p, miR-185, miR-22, and miR-26a in H517Q MNs. Intronic counts for the isogenic sample were set to 1. N = 2. Adjusted p value for PDE2A/miR-139 = 3.8e–10 estimated using DESeq2 analysis.
(H) Relative expression of pri-miRNAs for miR-139-5p, miR-185, miR-22, and miR-26a in sALS MNs. Intronic counts for the control MNs were set to 1. N = 13 for ALS and N = 8 for controls. N: number of independent differentiations. Error bars indicate SEMs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.