Figure 1. Targeting viral replication and downstream interferon signaling ameliorates chronic COVID-19.
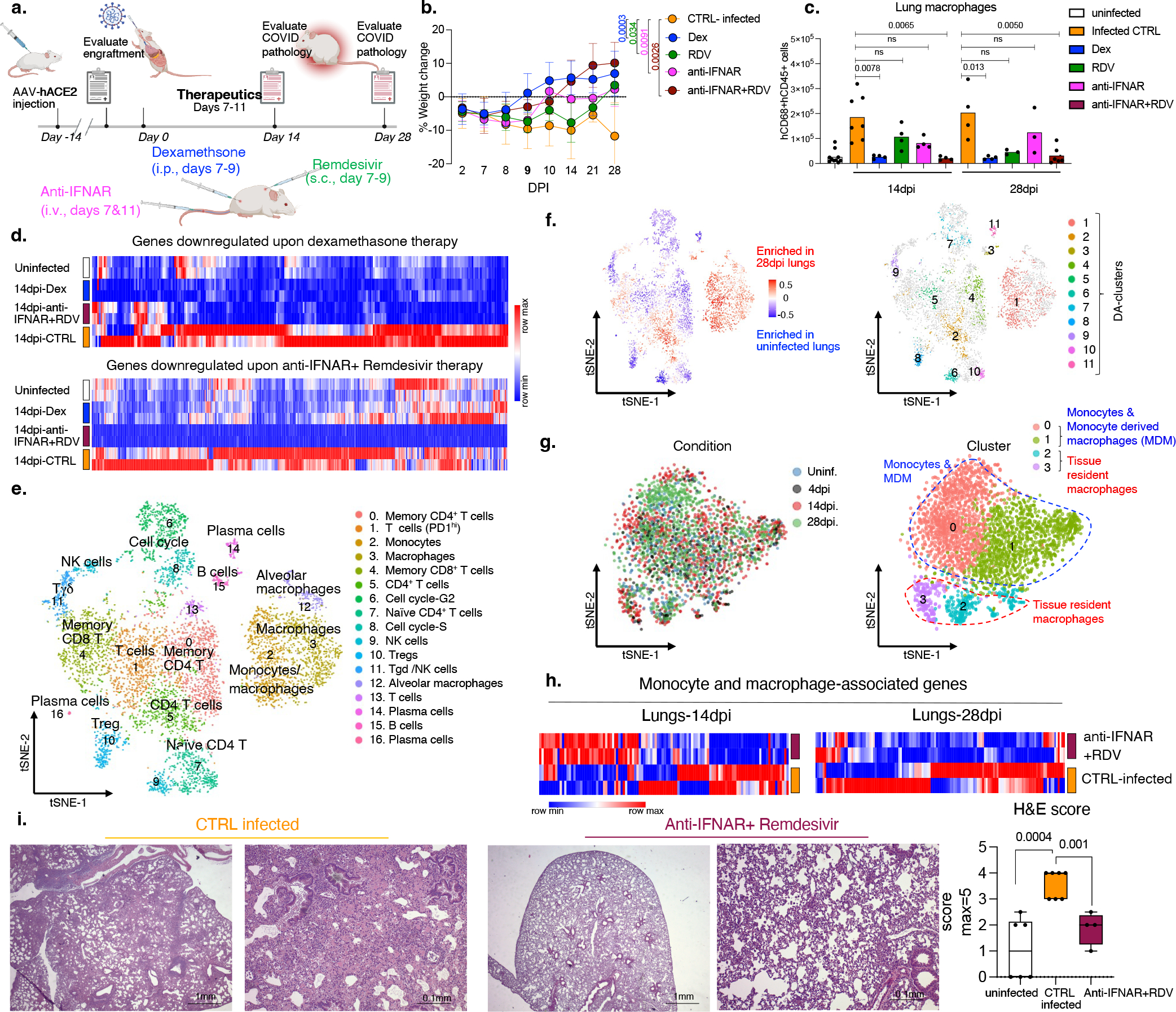
a. Therapy Schematic: SARS-CoV-2 infected MISTRG6-hACE2 mice were treated with Dexamethasone(dex) and Remdesivir(RDV) at 7,8,9 dpi, with anti-IFNAR2 at 7, 11dpi and analyzed at 14 or 28dpi.
b. Post-infection weight changes. 28dpi: CTRL-infected n=5, Dex, anti-IFNAR2+RDV n=4, RDV, anti-IFNAR2 n=3 mice examined over at least 2 experiments. Means with SD. Unpaired, two-tailed t-test.
c. Human macrophages in lungs. Uninfected: n=10; 14dpi: CTRL-infected n=7 Dex, RDV, anti-IFNAR, anti-IFNAR+RDV n=4; 28dpi: CTRL-infected, Dex n=4, RDV, anti-IFNAR n=3, anti-IFNAR+RDV n=6 mice examined over 3 experiments. Means with datapoints. Unpaired, two-tailed t-test.
d. Heatmap of genes suppressed by therapy in lungs (Log2, Foldchange >1; P-adj with the Bonferroni correction <0.05). Differential expression by DESeq2. Statistics by Wald-test. Transformed (min-max), normalized counts of duplicates. Hierarchical clustering (one-minus Pearson).
e. t-distributed stochastic neighbor embedding (t-SNE) plot of human immune cells from uninfected or infected lungs (28dpi). Pooled duplicates. Cluster marker genes identified with Wilcoxon-test (Extended data Fig. 2e). Uninfected=3,655, 28dpi=3,776 cells analyzed.
f. t-SNE plots highlighting differentially abundant (DA) human immune cell populations identified by DA-seq76. Top: Distribution/enrichment of DA-populations. Bottom: DA-clusters.
g. t-SNE plots of human monocyte/macrophage clusters from 4dpi, 14dpi and 28dpi and uninfected lungs. Left: dpi, right: clusters. Different conditions integrated as described in methods75. Marker genes identified with Wilcoxon-test (Extended data Fig.3a,b). P-adj with the Bonferroni correction. Uninfected=438, 4dpi=336, 14dpi=793, 28dpi=1368 cells analyzed.
h. Heatmap visualizing response to the combined therapy based on DEGs associated with monocytes and macrophages. Transformed (min-max), normalized expression of duplicates. Hierarchical clustering (one-minus Pearson).
i. Representative H&E staining and box plot of histopathological scores. Uninfected n=6, CTRL-infected n=7, anti-IFNAR2+RDV n=4 mice examined over 3 experiments. Whiskers: smallest/minimum to the largest/maximum value. Box: 25th-75th percentiles. Center line: median. Unpaired, two-tailed, t-test. Data associated with dexamethasone used here as a control have been reported19.
