Figure 2: Experimental components of SNV Recommended Track Set.
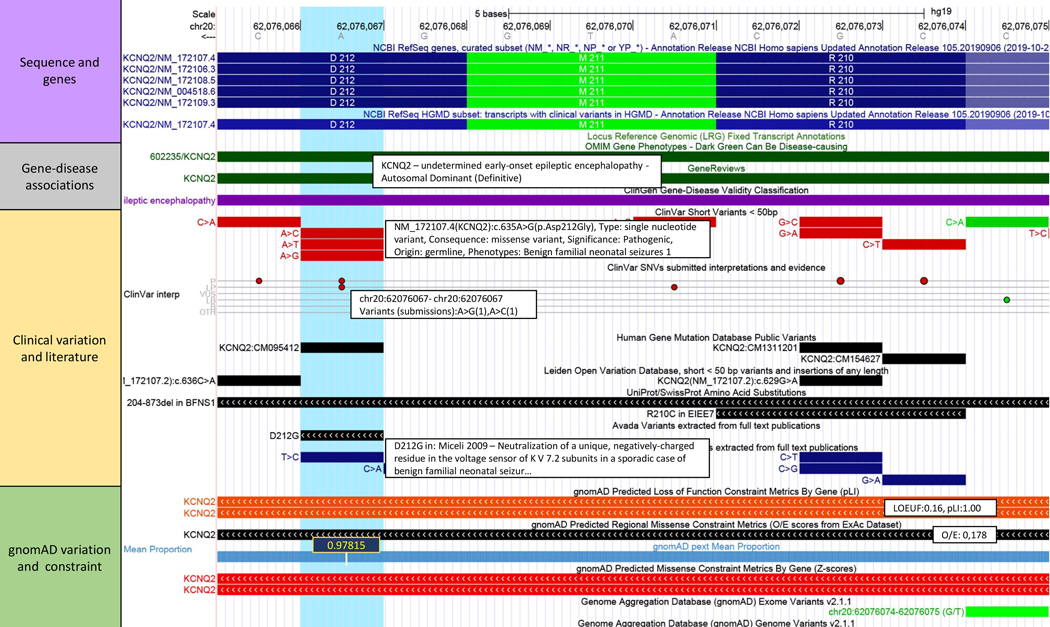
The variant record for NM_172107.4(KCNQ2):c.635A>G(p.Asp212Gly) (blue highlight) is shown, with UCSC Recommended Clinical SNVs track set applied: https://genome.ucsc.edu/s/abenet/HumanMutationFig2. This variant is located in the KCNQ2 gene. This gene has been ‘definitively’ associated with early-onset epileptic encephalopathy, an autosomal dominant inherited disease. The ClinVar Short Variants and SNVs tracks show that all three possible single nucleotide substitutions have been described at this position with pathogenic or likely pathogenic classifications. Each described variant leads to a missense change of the Asp residue. The same amino acid and nearby region is affected by mostly pathogenic variants. These and nearby variants show literature entries in the PubMed and HGMD database. The constraint metrics tracks show that the KCNQ2 gene has a high intolerance to both loss-of-function (LOEUF:0.16, pLI:1.00) and missense variation (Z:4.04), which is consistent with the number of classified pathogenic variants shown in this region of the gene. The pext score shows high isoform expression level across tissues (pext:0.97815). Other prediction data tracks are shown in Figure 3.
