Figure 3: Computational components of SNV Recommended Track Set.
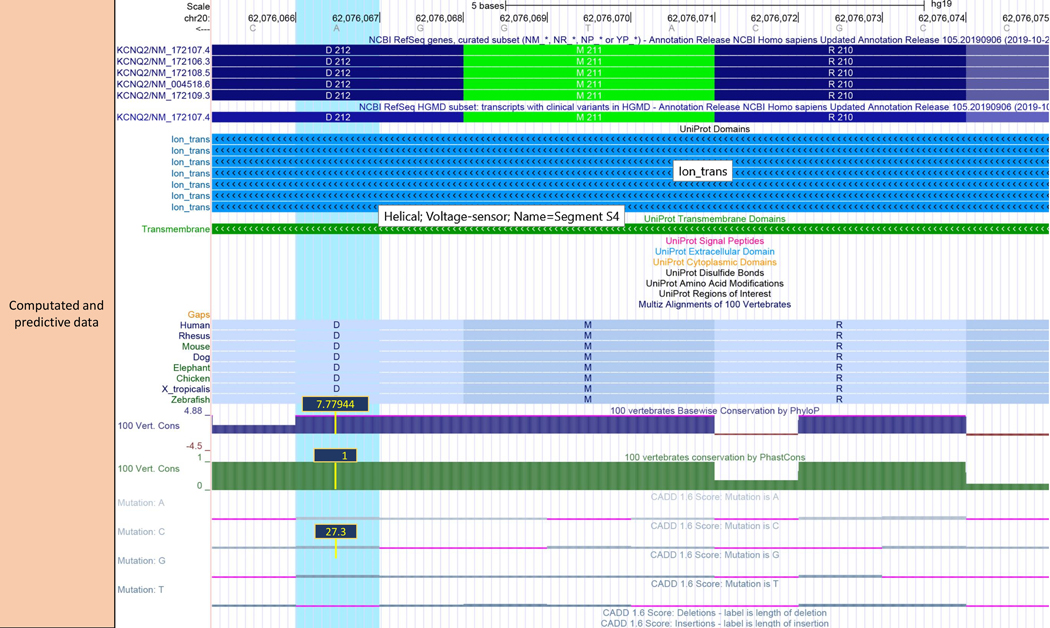
The UCSC Recommended Clinical SNVs computational and predictive tracks are shown for the variant NM_172107.4(KCNQ2):c.635A>G(p.Asp212Gly) (blue highlight): https://genome.ucsc.edu/s/abenet/HumanMutationFig3. The blue and green blocks of the Uniprot Annotations tracks at the top of the image show that the variant is located in a helical transmembrane domain with voltage-sensor function. Clicking on the green transmembrane item will show the details page where users can find more information about the function of this protein, which is a member of the potassium voltage-gated channel subfamily KQT, and outlinks to the Uniprot record. Mouseovers in PhyloP and PhastCons tracks show significant conservation scores at this nucleotide position (~1, 7.7) the aspartic acid residue which is fully conserved across all species of the Multiz 100-species alignment track. A CADD Phred score greater than 20 (C-score:27.3) (range 1 to 100) indicates that this substitution is predicted to be within the 1% most deleterious substitutions in the human genome. Taken together, the evidence extracted from these data suggest that this variant could have an inactivating effect on protein function. This is consistent with the lines of evidence from the clinical, population and constraint data, as described in Figure 2.
