Figure 4: Recommended Track Set for CNVs.
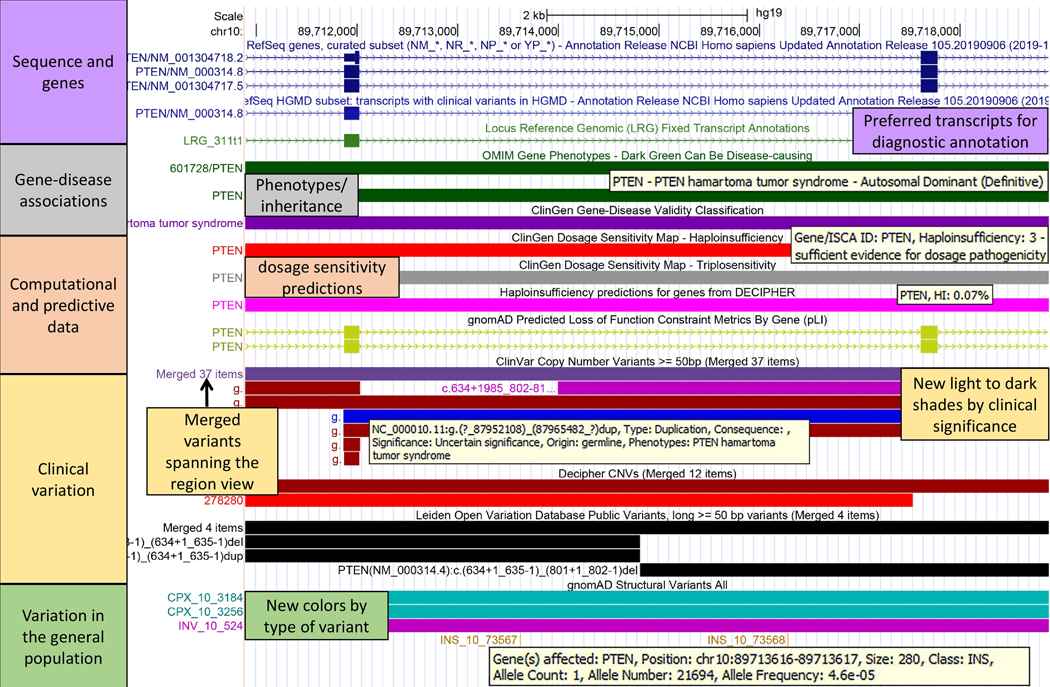
The gene record on hg19 for human PTEN exons 6 to 8 (NM_000314.8) is shown with UCSC Recommended Clinical CNVs track set applied: https://genome.ucsc.edu/s/abenet/HumanMutationFig4. The LRG and NCBI RefSeq HGMD tracks show the preferred transcript used for clinical annotation. The PTEN gene shows definitive association with the autosomal dominant PTEN hamartoma tumor syndrome. It is predicted to be an haploinsufficiency gene with sufficient evidence for dosage pathogenicity (ClinGen Dosage score: 3, DECIPHER HI: 0.07%). The clinical variation tracks show pathogenic deletions spanning various exons (dark red) and duplications of benign or unknown significance (light and middle blue). Mouseovers in the ClinVar track show CNV details about clinical significance and phenotypes described for that variant. Thirty-five variants in the ClinVar track and 12 variants in the DECIPHER track spanning the browser window are shown collapsed (Merged items), simplifying the view when zoomed to single-exon events. New colors for new types of variants are shown in the gnomAD Structural Variants track (green: complex, purple: inversion, orange: insertion) and mouseovers in this track show population counts and frequencies.
