Abstract
Rare disease diagnostics and disease gene discovery have been revolutionized by whole‐exome and genome sequencing but identifying the causative variant(s) from the millions in each individual remains challenging. The use of deep phenotyping of patients and reference genotype−phenotype knowledge, alongside variant data such as allele frequency, segregation, and predicted pathogenicity, has proved an effective strategy to tackle this issue. Here we review the numerous tools that have been developed to automate this approach and demonstrate the power of such an approach on several thousand diagnosed cases from the 100,000 Genomes Project. Finally, we discuss the challenges that need to be overcome if we are going to improve detection rates and help the majority of patients that still remain without a molecular diagnosis after state‐of‐the‐art genomic interpretation.
Keywords: diagnostics, phenotypes, rare disease, variant prioritization
1. INTRODUCTION
Rare diseases (RDs) are estimated to affect a substantial proportion of the population, estimated at 6% by one study although exact numbers vary considerably depending on definitions of RD, methodologies, and sources of data (Ferreira, 2019; Haendel et al., 2019). In addition, most RD patients undergo considerable medical odysseys before a diagnosis (Splinter et al., 2018). Next‐generation sequencing has started to transform RD diagnostics and research and numerous programs have demonstrated improved diagnostic yields from large‐scale whole‐exome and genome sequencing (WES and WGS) studies as well as efficient identification of novel disease−gene associations: Care4Rare (Dyment et al., 2015), Centers for Mendelian Genomics (Posey et al., 2019), Undiagnosed Diseases Network (Splinter et al., 2018). In particular, the UK 100,000 Genomes Project has transformed the way that genomics is used in the UK's National Health Service (NHS) for RDs with a WGS now the standard genetic test for many types of RD (Smedley et al., 2021).
Despite these successes, the causative mutations in the genomes of the majority of patients remain undetected after WES or WGS with diagnostics yields of 25%–50% (Clark et al., 2018; de Ligt et al., 2012; Rauch et al., 2012; Tammimies et al., 2015; Y. Yang et al., 2013, 2014; Zhu et al., 2015). Numerous variants in an affected individual typically remain after filtering a WES or WGS for variants using standard strategies for RD candidacy. These include identifying variants that: (i) are extremely rare according to population sequencing database such as GnomAD (Karczewski et al., 2020), (ii) segregate with disease in extended pedigrees, and (iii) are predicted to be pathogenic using in silico algorithms such as, for example, REVEL (Ioannidis et al., 2016), MVP (Qi et al., 2018), PolyPhen‐2 (Adzhubei et al., 2010), CADD (Kircher et al., 2014), MutationTaster (Schwarz et al., 2010), and SIFT (P. C. Ng & Henikoff, 2001). In high‐throughput and often under‐resourced healthcare settings, the causative variant can often be overlooked in this background. One increasingly adopted approach is to collect detailed clinical phenotype data on each affected individual using the Human Phenotype Ontology (HPO; Köhler et al., 2021) and compare that to reference phenotypic knowledge associated with each candidate variant and gene to narrow down the search further. The majority of the projects described above have successfully used this approach to improve diagnostic outcomes and many groups have built computational frameworks and pipelines to automate the phenotypic comparisons (summarized in Table 1).
Table 1.
Phenotype‐driven variant prioritization software
| Software | Low‐throughput web access | High‐throughput programmatic access | GRCh38 analysis | Family‐based analysis | SNV analysis | SV analysis | Noncoding analysis | Novel disease gene discovery through model organism, pathway, PPI data etc. | Natural language processing of latest literature | Reference data update (first published) |
|---|---|---|---|---|---|---|---|---|---|---|
| Exomiser framework (Smedley et al., 2015) including PhenIX (Zemojtel et al., 2014) and Genomiser (Smedley et al., 2016) | Yes | Yes | Yes | Yes | Yes | Yes | Yes (Genomiser only) | Yes | No | 2021 (2014) |
| AMELIE (Birgmeier et al., 2020) | Yes | Yes | No | Yes | Yes | No | No | No | Yes | 2021 (2020) |
| AnnotSV (Geoffroy et al., 2021) | Yes | Yes | Yes | No | Yes | Yes | Yes | No | No | 2021 (2018) |
| SvAnna (Danis et al., 2021a) | No | Yes | Yes | No | No | Yes | Yes | No | No | 2021 (2021) |
| LIRICAL (Robinson et al., 2020) | No | Yes | Yes | No | Yes | No | No | No | No | 2021 (2021) |
| xRare (Q. Li, Zhao, et al., 2019) | No | Yes | No | No | Yes | No | No | Yes | No | 2018 (2019) |
| VARPP (Anderson et al., 2019) | No | Yes | No | No | Yes | No | No | No | No | 2019 (2019) |
| DeepPVP (Boudellioua et al., 2019) | No | Yes | No | No | Yes | No | No | Yes | No | 2018 (2019) |
| MutationDistiller (Hombach et al., 2019) | Yes | No | No | No | Yes | No | No | Yes | No | 2018 (2019) |
| GenIO (Koile et al., 2018) | Yes | No | No | No | Yes | No | No | No | No | 2017 (2018) |
| wAnnovar (H. Yang & Wang, 2015) | Yes | No | Yes | No | Yes | No | No | Yes | No | 2017 (2015) |
| QueryOR (Bertoldi et al., 2017) | Yes | No | No | Yes | Yes | No | No | Yes | No | 2017 (2017) |
| BierApp (Alemán et al., 2014) | Yes | No | No | Yes | Yes | No | No | No | No | 2016 (2014) |
| OVA (Antanaviciute et al., 2015) | Yes | No | No | No | Yes | No | No | Yes | No | 2015 (2015) |
| Phen‐Gen (Javed et al., 2014) | No | Yes | No | Yes | Yes | No | Yes | Yes | No | 2013 (2014) |
| eXtasy (Sifrim et al., 2013) | Yes | Yes | No | No | Yes | No | No | Yes | Yes | 2013 (2013) |
Note: Peer‐reviewed, freely available (to academics/nonprofits at a minimum) software offering, HPO‐based prioritization of variants from rare disease case‐based VCF files. Software was reviewed for a range of features required for accurate, up‐to‐date interpretation at scale.
Abbreviations: SNV, single nucleotide variant; SV, structural variant.
A whole range of computational algorithms has been deployed in these tools incorporating natural language processing, machine learning, and artificial intelligence including deep neural networks, semantic similarity, and statistical probability approaches such as likelihood ratios. Each of the published tools also varies in terms of licensing, whether high‐throughput programmatic use is possible and whether they support features such as human genome assembly GRCh38 and family‐based analysis (Table 1). However, only a handful of tools, including Exomiser, AMELIE, and LIRICAL, show evidence of active maintenance with underlying databases updated since 2019. Caution should be exercised when using the other tools as any of the numerous, recently discovered new disease−gene associations will likely not be detectable. Further illustrating the problems with long‐term maintenance of academic software, many of the tools were no longer available at their published locations and are therefore not included in Table 1: PhenoPro (Z. Li, Zhang, et al., 2019), OMIMExplorer (James et al., 2016), Phenoxome (Wu et al., 2019).
In this article, we first explore how phenotype‐driven methods can improve diagnostic yields for RD using a large cohort of 4877 affected individuals who had received a molecular diagnosis (i.e., solved cases) from the 100,000 Genomes Project (Turnbull et al., 2018) and a set of 184 causative structural variants and corresponding phenotypic data curated from the literature. We then discuss some of the future challenges in the field that need to be overcome to address the overwhelming numbers of RD patients that still do not receive a molecular diagnosis after the current standard of care analysis of their WES and WGS samples.
2. CLINICAL PHENOTYPES ARE CRITICAL FOR AUTOMATED DETECTION OF RD DIAGNOSES
We explored the potential of phenotype‐driven variant prioritization software on 4877 molecularly diagnosed cases from the 100,000 Genome Project. This cohort represents diagnoses in some 1315 different genes for probands recruited under eligibility criteria for 257 broad clinical indications across all major branches of RD, for example, cardiovascular, ciliopathies, dermatological, dysmorphic and congenital abnormalities, endocrine, gastroenterological, growth, hematological, hearing, metabolic, neurology and neurodevelopmental, ophthalmological, renal and urinary tract, respiratory, rheumatological, skeletal, and finally tumor syndromes. Varying numbers of affected and unaffected family members were recruited and sequenced alongside the proband, bringing the total number of genomes analyzed in this cohort to 10,887. HPO terms were collected from the recruiting clinicians for each participant: median of 4 positive terms and range 1−61 per participant. Previous studies have shown that having more HPO terms per patient increases the chances of a diagnostic variant being ranked top by phenotype‐based, variant prioritization tools, but using more than five terms only improves performance slightly (Thompson et al., 2019).
To analyze this large cohort we required software that could be run on both GRCh37 and GRCh38 single nucleotide variant (SNV)/insertion‐deletion (indel) Variant Call Format (VCF) files (Danecek et al., 2011), offered local installation in the Genomics England research environment, as well as high‐throughput, programmatic querying. Exomiser and LIRICAL were the only two tools that satisfied these conditions and the performance of both is shown in Figure 1. Overall, Exomiser was able to prioritize 82.6%, 91.3%, 92.4%, and 93.6% of the 4877 diagnoses in the top, top 3, top 5, and top 10 ranked candidates. This demonstrates the effectiveness of a phenotype‐driven approach, across the whole breadth of RD, in automatically detecting the diagnostic variant(s) from the several million variants in the family WGS samples. Performance was similar for the more challenging singleton samples (N = 1591), demonstrating that sequencing of family members is not necessarily critical to identify a disease‐causative variant when deep, clinical phenotypes are collected. LIRICAL can currently only be run on singleton samples and, despite showing slightly reduced performance relative to Exomiser, still achieved efficient prioritization of diagnoses with 85.2% of diagnoses detected in the top 5 compared to 94.3% by Exomiser for these samples. Exomiser is able to use local frequency data available for the 100,000 Genomes Project to remove many false‐positive variant calls, which likely explains much of this difference in the recall. Where diagnoses were not recalled by the automated software, this was due to variants being filtered out as they were flagged as low quality in the VCF (1%), had unusually high minor allele frequencies (2%), or were incompletely penetrant (3%).
Figure 1.
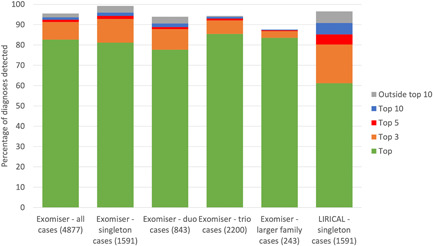
Recall of 4877 known molecular diagnoses from the 100,000 Genomes Project. Exomiser and LIRICAL were run using their standard settings and the percentage of molecular diagnoses detected as the top hit, in the top 3, 5, or 10 hits, or outside the top 10 are shown in the stacked bars. Performance for Exomiser was further broken down into whether the cases are singletons, duos (one parent sequenced), trios (both parents sequenced), or even larger family structures, for example, siblings sequenced as well.
Exomiser, like most of the methods described above, combines variant‐ and phenotype‐associated data into a single combined score or probability (Figure 2). The variant‐based filtering and scoring utilize minor allele frequencies from local and population sequencing sources, in silico predicted pathogenicity, variant molecular consequence for the gene, and segregation across affected and unaffected members. The phenotype‐based scoring is obtained from the semantic similarity between the proband's phenotype and the phenotypic profiles of human diseases and model organisms associated with the gene or nearby neighbors in a protein−protein interaction network.
Figure 2.
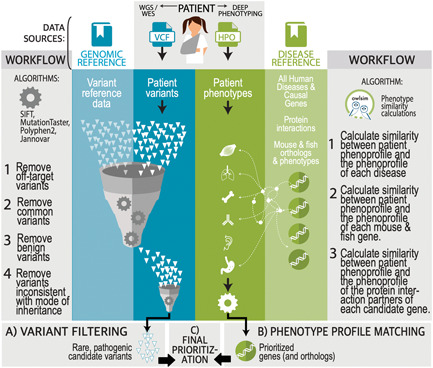
Representation of Exomiser's phenotype‐based prioritization strategy. Exomiser takes as input a set of clinical phenotypes encoded as HPO terms as well as a patient VCF file from WES/WGS sequencing followed by variant calling. Optionally, these VCF files can be multisample, representing the sequences of other affected and unaffected family members, and further data on the pedigree is also supplied. Under default settings, Exomiser then removes any variants that are not protein‐coding, above minor allele frequency thresholds of 0.1% for dominant and 2% recessive modes of inheritance, and that do not segregate with the disease (except well‐supported pathogenic/likely pathogenic ClinVar variants retained regardless of location or frequency). Remaining variants for each possible mode of inheritance are then scored based on the rarity of the variant, predicted consequence, and the output of in silico pathogenicity prediction algorithms such as REVEL, MVP, SIFT, PolyPhen‐2, and MutatationTaster. In parallel, existing phenotypic data for each gene associated with these candidate variants are compared to the patient phenotypes, a phenotype score calculated, and combined with the variant score to produce a final Exomiser score that can be used to rank the candidate variants. This phenotypic evidence comes from known disease associations (OMIM, Orphanet) and model organism databases (MGI, IMPC, ZFIN) as well as nearby gene neighbors in the StringDB protein−protein association network.
The importance of combining both variant and phenotypic data is seen in Figure 3a where the recall and precision for detecting the diagnoses in the 4877 are shown across the full range of Exomiser's variant, phenotype, and combined score cutoffs. Although a high recall (0.92) can be obtained using a variant score threshold of 0.8, the precision is poor (0.04) meaning an average of 25 variants per case have to be reviewed by a clinical geneticist before a report can be issued. In contrast, a phenotype score cutoff of 0.6 can be used to achieve a better precision (0.15) but with a considerably reduced recall (0.80). Combining both into the Exomiser score with a threshold of 0.7 allows accurate recall (0.89) with reasonable precision (0.15). Figure 3b summarizes how combining variant and phenotype data into an Exomiser score is critical for efficient variant prioritization with 82% of diagnoses recalled as the top hit compared to only 33% and 55% using the variant and phenotype scores respectively. In practice, we recommend users review the top 5 Exomiser candidates regardless of the score where a recall and precision of 0.92 and 0.18, respectively, can be obtained (Figure 3c).
Figure 3.
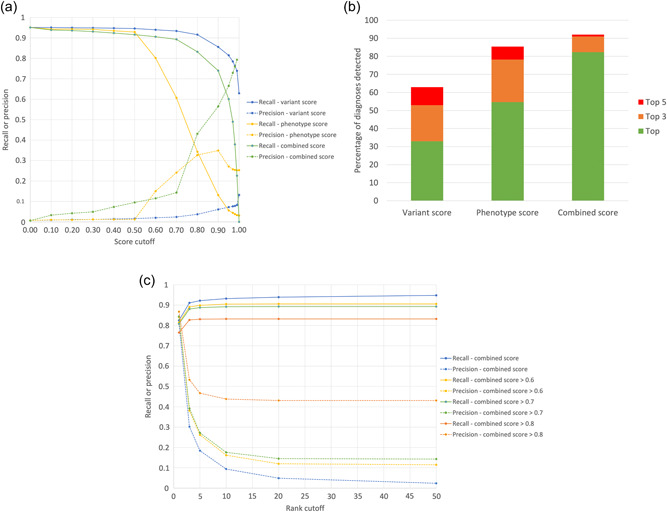
Exomiser performance on 4877 known molecular diagnoses from the 100,000 Genomes Project. (a) Recall and precision at different score thresholds for the variant, phenotype, or combined Exomiser score. (b) Percentage of diagnoses detected as the top hit or in the top 3 or 5 hits when ranking by variant, phenotype to combined Exomiser score. (c) Recall and precision when ranking by Exomiser combined score as well as additional score thresholds.
For many of the tools, the fact that they have not been upgraded to GRCh38 since publication prevented their evaluation here, and this is likely a reflection of the challenges of software maintenance in academia. Although it is difficult to predict the relative performance of the tools if they could be updated to work with the latest genome assembly and disease data, we do expect that, in general, all tools would demonstrate that a combined variant and phenotype‐based approach is highly effective.
3. CHALLENGES IN RD INTERPRETATION
Although most of the phenotype‐based variant prioritization methods have demonstrated impressive recall and precision on known molecular diagnoses, there still remain a proportion of those diagnoses that are not detected at all, for example, ~5% in the 100,000 Genomes Project benchmarking of Exomiser, as well as the much larger problem of most patients still not receiving a molecular diagnosis after a comprehensive analysis of their WES/WGS data, for example, 75% of 100,000 Genomes Project probands (Smedley et al., 2021). Better methods are needed to detect these missed diagnoses that improve: (i) the detection and prioritization of noncoding and structural variants, (ii) identify causative variants in genes that have not previously been associated with human disease, and (iii) deal with more complex genetic scenarios such as incomplete penetrance. Improvements are also required to allow easier reinterpretation of unsolved cases, simpler sharing of phenotype data, and diagnostics in a prenatal context. Here we will discuss the latest advances in these areas.
3.1. Prioritization of noncoding variants
A substantial proportion of RD diagnoses are likely to involve noncoding variants, for example, 4% of molecular diagnoses reported in the 100,000 Genomes Project pilot paper, demonstrating that WGS can accurately detect such diagnoses (Smedley et al., 2021). However, most pipelines are not routinely pursuing such diagnoses, largely due to the problem of overwhelming numbers of variants to interpret and validate. Phenotype‐based algorithms such as Genomiser (Smedley et al., 2016, part of the Exomiser framework) can automatically highlight candidate variants across the whole genome including enhancers, promoters, untranslated regions, and introns; previous benchmarking revealed that 77% of known noncoding molecular diagnoses could be recalled as the top candidate in WGS samples (Smedley et al., 2016). However, the issue of how to efficiently perform functional validation of novel noncoding variants limits the wider application of such approaches.
Researchers have therefore focussed their efforts on variants that change mRNA splicing as these are much more amenable to high‐throughput validation through techniques such as transcriptomics. The simplest definition of a splice variant includes variants that affect the most conserved AG/GT dinucleotides of the intron termini. However, variants at other splice site positions or variants located outside of the splice sites were also shown to cause defective splicing by introducing cryptic splice sites or by disrupting splicing regulatory element binding sites (Boichard et al., 2008). Recent algorithms such as SQUIRLS (Danis et al., 2021b) and SpliceAI (Jaganathan et al., 2019) have revolutionized the detection of such variants in WGS. Being able to integrate these new variant‐based algorithms into the phenotype‐based tools promises to deliver many additional diagnoses, as in some genes up to 50% of all disease‐causing variants are splice variants (Ars et al., 2000). Exomiser allows new variant deleteriousness or pathogenicity algorithms to be immediately incorporated into the analysis as tabix‐format score files. Initial exploration of this approach using SpliceAI and SQUIRLS on unsolved cases from the 100,000 Genomes Project has revealed tens of thousands of predicted pathogenic, cryptic splice variants within genes known to be associated with the patient's condition. It can be anticipated that intersecting these candidate variants with large‐scale transcriptomic analysis will allow the detection of many new molecular diagnoses. The direct integration of transcriptomic analysis into existing phenotype‐based variant prioritization software would also make this process much more efficient and powerful, building on existing gene prioritization approaches such as GADO (Deelen et al., 2019).
3.2. Prioritization of structural variants
Similarly, many unsolved RD cases are thought to involve structural variants (SVs), either alone or in combination with SNVs/indels. Even with the current limitations of calling SVs from short‐read WGS samples, 8% of the diagnoses reported by the 100,000 Genomes Project involved SVs (Smedley et al., 2021).
The challenge with SV prioritization ultimately stems from the primary technological challenges of sequencing, assembly, and calling of structural variants compared to short sequence variants, especially using short‐read technologies (Mahmoud et al., 2019). SV callers for both long‐ and short‐read technologies have varied performance depending on the class of SV they are calling, with insertions being a particularly troublesome class for reliable detection (Kosugi et al., 2019). In general, long‐read sequencing offers improved detection of SVs. However, while whole‐genome short‐read sequencing costs have dramatically reduced over the past decade, long‐read costs are still beyond what would be tolerated for routine diagnosis.
While the VCF specification has support for describing SVs, it is less well‐specified compared to sequence variants, with several open tickets (https://github.com/samtools/hts-specs/issues/544) under discussion for v4.4. Moreover, callers often follow the specification in an idiosyncratic manner, which makes it exceptionally difficult for variant prioritization software to reliably utilize the calls. One of the most powerful metrics for judging variant pathogenicity is variant frequency where pathogenic variants are often absent or present at very low frequencies in databases such as gnomAD. GnomAD‐SV (Collins et al., 2020) now offers a reference database produced from high‐coverage sequencing to perform this task also for SVs. However, the recent gnomAD‐SV data set was created from 14,891 individuals and is far smaller than the original gnomAD SNV/indel data set with around 140,000 individuals. This reduces the filtering power based on variant frequency of gnomAD‐SV. Other resources such as DECIPHER, DGV, and dbVAR also contain SVs but data such as SV type, insertion length, and copy number are not always recorded consistently within or between resources. A further problem with trying to utilize these resources is that SVs are harder to categorize, far longer, and often have imprecise boundaries when compared to small variants, and are therefore significantly harder to look up in reference databases. Guidelines for reporting clinical pathogenicity of structural variants have only been introduced recently (Riggs et al., 2020) compared to the long‐established ones for SNV variants (Richards et al., 2015), leading to fewer high‐quality clinical assertions in ClinVar (Landrum et al., 2018) for tools to reference.
Despite these challenges, several phenotype‐based prioritization tools have recently emerged that offer SV prioritization. SvAnna (Danis et al., 2021a) focuses on SVs called from long‐read technologies. AnnotSV (Geoffroy et al., 2021) in contrast has a short‐read focus. The latest release of Exomiser (13.0.0) allows phenotype‐based prioritization of SVs alongside SNVs/indels so that the impact of the SVs on the coding regions of one or more genes is assessed alongside any rare, predicted damaging SNVs/indels present under various segregation models for each affected individual.
The ability of Exomiser to prioritize known molecular diagnoses involving an SV is shown in Figure 4. Previously described phenopackets (https://phenopacket-schema.readthedocs.io/en/latest/index.html) representing curated phenotypic and pedigree data from the literature (Danis et al., 2021a) were used as input to Exomiser alongside corresponding VCFs containing the curated variant(s) added to a control WGS VCF file based on either short‐ or long‐read technologies. The former used an Illumina short‐read sample with SVs called using Manta and Canvas. For the long‐read benchmarking we used a Genome in a Bottle (GIAB) sample generated by PacBio sequencing and pbsv calling (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/PacBio_pbsv_05212019/HG002_GRCh38.pbsv.vcf). Exomiser was able to prioritize 74% of the SV diagnoses as the top‐ranked candidate and 89% in the top 5 for the short‐read samples. The long‐read samples were more problematic with performance dropping to 61% and 78% for the top and top 5 ranked candidates, but still showing relatively effective prioritization of SV diagnoses. Only 14 SV diagnoses were completely missed by Exomiser: nine involving SVs that disrupt noncoding regions which are not currently handled and five unspecified breakend (BND)‐type SVs that again are not currently supported. Twenty‐eight of the 184 curated known SV diagnoses involve an SNV/indel in compound heterozygosity with an SV and in all cases Exomiser was able to detect both variants and prioritize the diagnosis effectively in the top 3 ranked candidates. The same phenopackets have already been assessed for SvAnna and AnnotSV using a different set of long‐read, pbsv called VCFs and showed 61% and 86% in the top and top 5 candidates for SvAnna and 60% and 65% for AnnotSV (Danis et al., 2021a).
Figure 4.
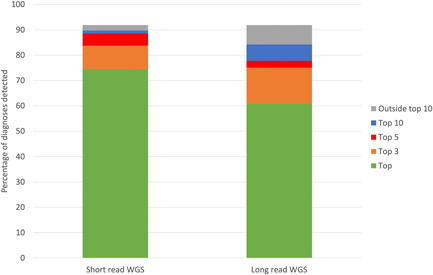
Exomiser recall of 184 known SV diagnoses described in the literature. Previously described phenopackets representing known SV diagnoses curated from the literature were used as input to Exomiser along with short‐ or long‐read‐based SV VCF files. Exomiser was run using standard settings and the percentage of diagnoses detected as the top hit, in the top 3, 5, or 10 hits, or outside the top 10 are shown in the stacked bars.
3.3. Incomplete penetrance
Exomiser and Phen‐Gen are the only phenotype‐based variant prioritization tools to offer the option of allowing for incomplete penetrance. In Exomiser 13.0.0, the user can configure the analysis to retain variants in unaffected family members instead of removing them as part of the standard filtering pipeline. Phen‐Gen has a stringency setting to adjust the level of penetrance. Allowing for incomplete penetrance obviously leads to more candidates to review per case. Both Exomiser and Phen‐Gen apply these settings across the genome and future improvements to restrict to a curated set of genes with known incomplete penetrance would reduce the number of candidates and improve performance. We were able to benchmark Exomiser on 35 families from the 100,000 Genomes Project with incompletely penetrant molecular diagnoses and show that 54% were still detected as the top‐ranked candidate, 77% in the top 3 with a further 14% found outside the top 10. Phen‐Gen benchmarking was not possible on these samples as GRCh38 analysis is not enabled. Without accounting for the incomplete penetrance, none of these diagnoses would have been detected.
3.4. Novel disease−gene discovery through phenotype‐based methods
The usual route for disease gene discovery involves identifying pathogenic/likely pathogenic variants in the same gene in several unrelated families with the same phenotype and then performing functional validation. The phenotype‐based tools that incorporate model organism data, pathway, and/or protein‐protein network approaches can prioritize variants in genes that have not previously been associated with human disease and potentially support this functional validation step. This is enabled by making use of existing knowledge, for example, from large‐scale efforts such as the International Mouse Phenotyping Consortium (Lloyd et al., 2020) that are characterizing the function of every protein‐coding gene through systematic mouse knockouts and phenotyping. For example, the Children's Hospital Los Angeles demonstrated the successful discovery of diagnoses in novel disease genes using a semi‐automated pipeline involving Exomiser (Ji et al., 2019). In another example, ANKRD17 was identified as a candidate gene for cases of intellectual disability in the 100,000 Genomes Project through the identification of highly ranked Exomiser candidate de novo variants based on protein−protein interaction evidence. Subsequent identification of further cases worldwide and functional characterization have now confirmed this association (Chopra et al., 2021).
3.5. Reinterpretation of unsolved cases
The power of reinterpretation to diagnose previously unsolved cases based on new knowledge of disease−gene associations has been widely reported in the last few years (Matalonga et al., 2021). Phenotype‐based, variant prioritization tools offer an ideal mechanism to implement efficient reinterpretation as long as the software is fast, easy to run, and, most critically, kept up‐to‐date. As we discussed at the beginning, only a few academic tools fulfill these criteria and a number of commercial solutions have emerged with similar offerings. However, the free, open‐source tools such as AMELIE, LIRICAL and Exomiser that do offer up‐to‐date reinterpretation have an attractive range of features:
-
(i)
programmatic access allowing high‐throughput analysis and, in the case of Exomiser and LIRICAL, simple, local installation so security around data transfer is not an issue (local reinstallation of latest versions required though). AMELIE offers the advantage of natural language processing of the latest literature to identify reference genotype to phenotype knowledge. Other tools such as Exomiser and LIRICAL rely on the curation of the latest disease−gene associations by OMIM and Orphanet and the associated phenotypes by the HPO team before this knowledge is available to the software;
-
(ii)
simple configuration, including sensible presets for exome‐ or genome‐based analysis in the case of Exomiser, so only the bare minimum of patient‐level information needs to be entered;
-
(iii)
standardized input using VCF files, HPO terms, and, in the case of Exomiser and LIRICAL, compatibility with the Global Alliance for Genomics and Health (GA4GH) approved Phenopacket standard that will allow future direct connection to electronic health record (EHR) systems;
-
(iv)
fast run times (<30 s for a WES, <5 min for a WGS) making regular reinterpretation feasible;
-
(v)
JavaScript Object Notation (JSON) output for incorporation into bioinformatics pipelines and, in the case of Exomiser and LIRICAL, user‐friendly HyperText Markup Language (HTML) output.
3.6. Standardized phenotype representation
Phenotype‐driven RD genome analysis tools have benefited enormously from standardized formats for capturing genomic variation from next‐generation sequencing technologies (VCF), yet until recently had no analog for describing patient phenotype. While the Human Phenotype Ontology (Köhler et al., 2021) has become the accepted standard for capturing patient phenotype from deep‐phenotyping for use in analysis, there is no standardized way of conveying this information to bioinformatics tools. Most tools rely on a simple list of phenotype terms or a disease identifier (e.g., from OMIM (Amberger et al., 2019), Orphanet (Pavan et al., 2017), or MONDO (http://obofoundry.org/ontology/mondo)) to try and convey this information, but this method cannot convey a complete description of an individual's phenotype including modifiers such as severity, laterality, and age of onset for each phenotype as well as their progression over time. The GA4GH Phenopacket (https://phenopacket-schema.readthedocs.io/en/v2/) aims to solve this by providing a standardized, structured format for describing patient‐level phenotypic features, allowing for a rich description of each feature including the absence, severity and time of onset. Since its initial release, several tools (LIRICAL, SvAnna, Exomiser, Phen2Gene (Zhao et al., 2020)) support the standard which offers significantly increased portability of phenotype data between these tools.
3.7. Interpretation of prenatal cases
While becoming routine for pediatric and adult diagnosis, the use of phenotype‐driven RD analysis for prenatal diagnosis is a developing area. Currently, the HPO has 151 terms in the subhierarchy starting from Abnormality of prenatal development or birth [HP:0001197]. Out of the 199,197 annotations to 7902 Mendelian diseases currently present in the HPO, roughly 0.5% refer to terms from the subhierarchy HP:0001197, such as Fetal distress [HP:0025116] or Short fetal humerus length [HP:0011429]. However, current knowledge of the prenatal manifestations of Mendelian disease remains limited. Prenatal genomic testing is becoming increasingly common for fetuses with suspected Mendelian disease but the interpretation of expanded prenatal sequencing is reliant on deeper fetal phenotyping (Gray et al., 2019). The HPO project is currently conducting a series of workshops in this area to expand the depth and breadth of relevant coverage.
4. CONCLUSIONS
WES and WGS are now widely used in both diagnostic and research settings. A large driver for the successful adoption of these strategies has been the collection of deep phenotype data using HPO terms and software allowing automated prioritization of variants. Without these tools, clinicians and researchers would be overwhelmed, in most cases, by the sheer number of candidate variants to interpret. Phenotype‐driven, RD diagnosis is now a part of routine clinical practice in the United Kingdom and many other healthcare systems. There are still numerous challenges to overcome before we can efficiently deliver on the promise of genomics to fully transform the diagnosis and eventual treatment of RD. Further development and adoption of standards are needed to connect EHR systems and the variant prioritization tools. More research and development of the tools are needed to identify the overlooked molecular diagnoses that are present in existing genomic samples as well as those that will emerge through further advances in omics technologies. However, we have come to a remarkable distance in the last decade since the first reported WES successes (S. B. Ng et al., 2010), and we expect considerable advances in all these areas in the next few years.
CONFLICTS OF INTEREST
Julius Jacobsen and Damian Smedley declare they previously acted as part‐time consultants for Congenica Ltd. The other authors declare no other potential conflicts of interest.
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH) grants 1R24OD011883, U54 HG006370, and NIH, National Institute of Child Health and Human Development 1R01HD103805‐01. This study was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support.
Jacobsen, J. O. B. , Kelly, C. , Cipriani, V. , Genomics England Research Consortium , Mungall, C. J. , Reese, J. , Danis, D. , Robinson, P. N. , & Smedley, D. (2022). Phenotype‐driven approaches to enhance variant prioritization and diagnosis of rare disease. Human Mutation, 43, 1071–1081. 10.1002/humu.24380
DATA AVAILABILITY STATEMENT
All data described in the paper are already provided in the paper except for access to the 100,000 Genomes Project samples which is by application to Genomics England.
REFERENCES
- Adzhubei, I. A. , Schmidt, S. , Peshkin, L. , Ramensky, V. E. , Gerasimova, A. , Bork, P. , Kondrashov, A. S. , & Sunyaev, S. R. (2010). A method and server for predicting damaging missense mutations. Nature Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemán, A. , Garcia‐Garcia, F. , Salavert, F. , Medina, I. , & Dopazo, J. (2014). A web‐based interactive framework to assist in the prioritization of disease candidate genes in whole‐exome sequencing studies. Nucleic Acids Research, 42, W88–W93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger, J. S. , Bocchini, C. A. , Scott, A. F. , & Hamosh, A. (2019). OMIM.org: Leveraging knowledge across phenotype−gene relationships. Nucleic Acids Research, 47, D1038–D1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D. , Baynam, G. , Blackwell, J. M. , & Lassmann, T. (2019). Personalised analytics for rare disease diagnostics. Nature Communications, 10, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antanaviciute, A. , Watson, C. M. , Harrison, S. M. , Lascelles, C. , Crinnion, L. , Markham, A. F. , Bonthron, D. T. , & Carr, I. M. (2015). OVA: Integrating molecular and physical phenotype data from multiple biomedical domain ontologies with variant filtering for enhanced variant prioritization. Bioinformatics, 31, 3822–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ars, E. , Serra, E. , García, J. , Kruyer, H. , Gaona, A. , Lázaro, C. , & Estivill, X. (2000). Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Human Molecular Genetics, 9, 237–247. [DOI] [PubMed] [Google Scholar]
- Bertoldi, L. , Forcato, C. , Vitulo, N. , Birolo, G. , De Pascale, F. , Feltrin, E. , Schiavon, R. , Anglani, F. , Negrisolo, S. , Zanetti, A. , D'Avanzo, F. , Tomanin, R. , Faulkner, G. , Vezzi, A. , & Valle, G. (2017). QueryOR: A comprehensive web platform for genetic variant analysis and prioritization. BMC Bioinformatics, 18, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgmeier, J. , Haeussler, M. , Deisseroth, C. A. , Steinberg, E. H. , Jagadeesh, K. A. , Ratner, A. J. , Guturu, H. , Wenger, A. M. , Diekhans, M. E. , Stenson, P. D. , Cooper, D. N. , Ré, C. , Beggs, A. H. , Bernstein, J. A. , & Bejerano, G. (2020). AMELIE speeds Mendelian diagnosis by matching patient phenotype and genotype to primary literature. Science Translational Medicine, 12, eaau9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boichard, A. , Venet, L. , Naas, T. , Boutron, A. , Chevret, L. , Baulny H. O. de, De Lonlay, P. , Legrand, A. , Nordman, P. , & Brivet, M. (2008). Two silent substitutions in the PDHA1 gene cause exon 5 skipping by disruption of a putative exonic splicing enhancer. Molecular Genetics and Metabolism, 93, 323–330. [DOI] [PubMed] [Google Scholar]
- Boudellioua, I. , Kulmanov, M. , Schofield, P. N. , Gkoutos, G. V. , & Hoehndorf, R. (2019). DeepPVP: Phenotype‐based prioritization of causative variants using deep learning. BMC Bioinformatics, 20, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, M. , McEntagart, M. , Clayton‐Smith, J. , Platzer, K. , Shukla, A. , Girisha, K. M. , Kaur, A. , Kaur, P. , Pfundt, R. , Veenstra‐Knol, H. , Mancini, G. M. S. , Cappuccio, G. , Brunetti‐Pierri, N. , Kortüm, F. , Hempel, M. , Denecke, J. , Lehman, A. , Causes, S. , Kleefstra, T. , … Gordon, C. T. (2021). Heterozygous ANKRD17 loss‐of‐function variants cause a syndrome with intellectual disability, speech delay, and dysmorphism. American Journal of Human Genetics, 108, 1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, M. M. , Stark, Z. , Farnaes, L. , Tan, T. Y. , White, S. M. , Dimmock, D. , & Kingsmore, S. F. (2018). Meta‐analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genomic Medicine, 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. L. , Brand, H. , Karczewski, K. J. , Zhao, X. , Alföldi, J. , Francioli, L. C. , Khera, A. V. , Lowther, C. , Gauthier, L. D. , Wang, H. , Watts, N. A. , Solomonson, M. , O'Donnell‐Luria, A. , Baumann, A. , Munshi, R. , Walker, M. , Whelan, C. W. , Huang, Y. , Brookings, T. , … Talkowski, M. E. (2020). A structural variation reference for medical and population genetics. Nature, 581, 444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P. , Auton, A. , Abecasis, G. , Albers, C. A. , Banks, E. , DePristo, M. A. , Handsaker, R. E. , Lunter, G. , Marth, G. T. , Sherry, S. T. , McVean, G. , & Durbin, R. (2011). The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis, D. , Jacobsen, J. O. B. , Balachandran, P. , Zhu, Q. , Yilmaz, F. , Reese, J. , Haimel, M. , Lyon, G. J. , Helbig, I. , Mungall, C. J. , Beck, C. , & Lee, C. (2021a). SvAnna: Efficient and accurate pathogenicity prediction for coding and regulatory structural variants in long‐read genome sequencing. bioRxiv 2021.07.14.452267. [DOI] [PMC free article] [PubMed]
- Danis, D. , Jacobsen, J. O. B. , Carmody, L. C. , Gargano, M. A. , McMurry, J. A. , Hegde, A. , Haendel, M. A. , Valentini, G. , Smedley, D. , & Robinson, P. N. (2021b). Interpretable prioritization of splice variants in diagnostic next‐generation sequencing. American Journal of Human Genetics, 108, 1564–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen, P. , van Dam, S. , Herkert, J. C. , Karjalainen, J. M. , Brugge, H. , Abbott, K. M. , van Diemen, C. C. , van der Zwaag, P. A. , Gerkes, E. H. , Zonneveld‐Huijssoon, E. , Boer‐Bergsma, J. J. , Folkertsma, P. , Gillett, T. , van der Velde, K. J. , Kanninga, R. , van den Akker, P. C. , Jan, S. Z. , Hoorntje, E. T. , Te Rijdt, W. P. , … Franke, L. (2019). Improving the diagnostic yield of exome‐ sequencing by predicting gene‐phenotype associations using large‐scale gene expression analysis. Nature Communications, 10, 2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment, D. A. , Tétreault, M. , Beaulieu, C. L. , Hartley, T. , Ferreira, P. , Chardon, J. W. , Marcadier, J. , Sawyer, S. L. , Mosca, S. J. , Innes, A. M. , Parboosingh, J. S. , Bulman, D. E. , Schwartzentruber, J. , Majewski, J. , Tarnopolsky, M. , Boycott, K. M. , FORGE Canada, C . (2015). Whole‐exome sequencing broadens the phenotypic spectrum of rare pediatric epilepsy: A retrospective study. Clinical Genetics, 88, 34–40. [DOI] [PubMed] [Google Scholar]
- Ferreira, C. R. (2019). The burden of rare diseases. American Journal of Medical Genetics. Part A, 179, 885–892. [DOI] [PubMed] [Google Scholar]
- Geoffroy, V. , Guignard, T. , Kress, A. , Gaillard, J.‐B. , Solli‐Nowlan, T. , Schalk, A. , Gatinois, V. , Dollfus, H. , Scheidecker, S. , & Muller, J. (2021). AnnotSV and knotAnnotSV: A web server for human structural variations annotations, ranking and analysis. Nucleic Acids Research, 49, W21–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, K. J. , Wilkins‐Haug, L. E. , Herrig, N. J. , & Vora, N. L. (2019). Fetal phenotypes emerge as genetic technologies become robust. Prenatal Diagnosis, 39, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendel, M. , Vasilevsky, N. , Unni, D. , Bologa, C. , Harris, N. , Rehm, H. , Hamosh, A. , Baynam, G. , Groza, T. , McMurry, J. , Dawkins, H. , Rath, A. , Thaxon, C. , Bocci, G. , Joachimiak, M. P. , Köhler, S. , Robinson, P. N. , Mungall, C. , & Oprea, T. I. (2019). How many rare diseases are there? Nature Reviews. Drug Discovery, 19, 77–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach, D. , Schuelke, M. , Knierim, E. , Ehmke, N. , Schwarz, J. M. , Fischer‐Zirnsak, B. , & Seelow, D. (2019). MutationDistiller: User‐driven identification of pathogenic DNA variants. Nucleic Acids Research, 47, W114–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis, N. M. , Rothstein, J. H. , Pejaver, V. , Middha, S. , McDonnell, S. K. , Baheti, S. , Musolf, A. , Li, Q. , Holzinger, E. , Karyadi, D. , Cannon‐Albright, L. A. , Teerlink, C. C. , Stanford, J. L. , Isaacs, W. B. , Xu, J. , Cooney, K. A. , Lange, E. M. , Schleutker, J. , Carpten, J. D. , … Sieh, W. (2016). REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. American Journal of Human Genetics, 99, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan, K. , Kyriazopoulou Panagiotopoulou, S. , McRae, J. F. , Darbandi, S. F. , Knowles, D. , Li, Y. I. , Kosmicki, J. A. , Arbelaez, J. , Cui, W. , Schwartz, G. B. , Chow, E. D. , Kanterakis, E. , Gao, H. , Kia, A. , Batzoglou, S. , Sanders, S. J. , & Farh, K. K. (2019). Predicting splicing from primary sequence with deep learning. Cell, 176, 535–548.e24. [DOI] [PubMed] [Google Scholar]
- James, R. A. , Campbell, I. M. , Chen, E. S. , Boone, P. M. , Rao, M. A. , Bainbridge, M. N. , Lupski, J. R. , Yang, Y. , Eng, C. M. , Posey, J. E. , & Shaw, C. A. (2016). A visual and curatorial approach to clinical variant prioritization and disease gene discovery in genome‐wide diagnostics. Genome Medicine, 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed, A. , Agrawal, S. , & Ng, P. C. (2014). Phen‐Gen: Combining phenotype and genotype to analyze rare disorders. Nature Methods, 11, 935–937. [DOI] [PubMed] [Google Scholar]
- Ji, J. , Shen, L. , Bootwalla, M. , Quindipan, C. , Tatarinova, T. , Maglinte, D. T. , Buckley, J. , Raca, G. , Saitta, S. C. , Biegel, J. A. , & Gai, X. (2019). A semiautomated whole‐exome sequencing workflow leads to increased diagnostic yield and identification of novel candidate variants. Cold Spring Harbor Molecular Case Studies, 5, a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski, K. J. , Francioli, L. C. , Tiao, G. , Cummings, B. B. , Alföldi, J. , Wang, Q. , Collins, R. L. , Laricchia, K. M. , Ganna, A. , Birnbaum, D. P. , Gauthier, L. D. , Brand, H. , Solomonson, M. , Watts, N. A. , Rhodes, D. , Singer‐Berk, M. , England, E. M. , Seaby, E. G. , Kosmicki, J. A. , … MacArthur, D. G. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, M. , Witten, D. M. , Jain, P. , O'Roak, B. J. , Cooper, G. M. , & Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics, 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, S. , Gargano, M. , Matentzoglu, N. , Carmody, L. C. , Lewis‐Smith, D. , Vasilevsky, N. A. , Danis, D. , Balagura, G. , Baynam, G. , Brower, A. M. , Callahan, T. J. , Chute, C. G. , Est, J. L. , Galer, P. D. , Ganesan, S. , Griese, M. , Haimel, M. , Pazmandi, J. , Hanauer, M. , … Robinson, P. N. (2021). The human phenotype ontology in 2021. Nucleic Acids Research, 49, D1207–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koile, D. , Cordoba, M. , Sousa Serro, M. de , Kauffman, M. A. , & Yankilevich, P. (2018). GenIO: A phenotype−genotype analysis web server for clinical genomics of rare diseases. BMC Bioinformatics, 19, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S. , Momozawa, Y. , Liu, X. , Terao, C. , Kubo, M. , & Kamatani, Y. (2019). Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biology, 20, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum, M. J. , Lee, J. M. , Benson, M. , Brown, G. R. , Chao, C. , Chitipiralla, S. , Gu, B. , Hart, J. , Hoffman, D. , Jang, W. , Karapetyan, K. , Katz, K. , Liu, C. , Maddipatla, Z. , Malheiro, A. , McDaniel, K. , Ovetsky, M. , Riley, G. , Zhou, G. , … Maglott, D. R. (2018). ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Research, 46, D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Zhao, K. , Bustamante, C. D. , Ma, X. , & Wong, W. H. (2019). Xrare: A machine learning method jointly modeling phenotypes and genetic evidence for rare disease diagnosis. Genetics in Medicine, 21, 2126–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Zhang, F. , Wang, Y. , Qiu, Y. , Wu, Y. , Lu, Y. , Yang, L. , Qu, W. J. , Wang, H. , Zhou, W. , & Tian, W. (2019). PhenoPro: A novel toolkit for assisting in the diagnosis of Mendelian disease. Bioinformatics, 35, 3559–3566. [DOI] [PubMed] [Google Scholar]
- de Ligt, J. , Willemsen, M. H. , van Bon, B. W. , Kleefstra, T. , Yntema, H. G. , Kroes, T. , Vulto‐van Silfhout, A. T. , Koolen, D. A. , de Vries, P. , Gilissen, C. , del Rosario, M. , Hoischen, A. , Scheffer, H. , de Vries, B. B. , Brunner, H. G. , Veltman, J. A. , & Vissers, L. E. (2012). Diagnostic exome sequencing in persons with severe intellectual disability. New England Journal of Medicine, 367, 1921–1929. [DOI] [PubMed] [Google Scholar]
- Lloyd, K. C. K. , Adams, D. J. , Baynam, G. , Beaudet, A. L. , Bosch, F. , Boycott, K. M. , Braun, R. E. , Caulfield, M. , Cohn, R. , Dickinson, M. E. , Dobbie, M. S. , Flenniken, A. M. , Flicek, P. , Galande, S. , Gao, X. , Grobler, A. , Heaney, J. D. , Herault, Y. , de Angelis, M. H. , … Brown, S. (2020). The Deep Genome Project. Genome Biology, 21, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud, M. , Gobet, N. , Cruz‐Dávalos, D. I. , Mounier, N. , Dessimoz, C. , & Sedlazeck, F. J. (2019). Structural variant calling: The long and the short of it. Genome Biology, 20, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalonga, L. , Hernández‐Ferrer, C. , Piscia, D. , Solve‐RD SNV‐indel working, g , Schüle, R. , Synofzik, M. , Töpf, A. , Vissers, L. E. L. M. , de Voer, R. , Solve‐Rd, D. , Solve‐Rd, D. , Solve‐Rd, D. , Solve‐Rd, D. , Tonda, R. , Laurie, S. , Fernandez‐Callejo, M. , Picó, D. , Garcia‐Linares, C. , Papakonstantinou, A. , … Solve‐RD, C. (2021). Solving patients with rare diseases through programmatic reanalysis of genome‐phenome data. European Journal of Human Genetics, 29, 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P. C. , & Henikoff, S. (2001). Predicting deleterious amino acid substitutions. Genome Research, 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, S. B. , Buckingham, K. J. , Lee, C. , Bigham, A. W. , Tabor, H. K. , Dent, K. M. , Huff, C. D. , Shannon, P. T. , Jabs, E. W. , Nickerson, D. A. , Shendure, J. , & Bamshad, M. J. (2010). Exome sequencing identifies the cause of a mendelian disorder. Nature Genetics, 42, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan, S. , Rommel, K. , Mateo Marquina, M. E. , Höhn, S. , Lanneau, V. , & Rath, A. (2017). Clinical practice guidelines for rare diseases: The Orphanet Database. PLoS ONE, 12, e0170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey, J. E. , O'donnell‐Luria, A. H. , Chong, J. X. , Harel, T. , Jhangiani, S. N. , Coban Akdemir, Z. H. , Buyske, S. , Pehlivan, D. , Carvalho, C. M. B. , Baxter, S. , Sobreira, N. , Liu, P. , Wu, N. , Rosenfeld, J. A. , Kumar, S. , Avramopoulos, D. , White, J. J. , Doheny, K. F. , Witmer, P. D. , … Centers for Mendelian, G . (2019). Insights into genetics, human biology and disease gleaned from family based genomic studies. Genetics in Medicine, 21, 798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, H. , Chen, C. , Zhang, H. , Long, J. J. , Chung, W. K. , Guan, Y. , & Shen, Y. (2018). MVP: Predicting pathogenicity of missense variants by deep learning. bioXriv.
- Rauch, A. , Wieczorek, D. , Graf, E. , Wieland, T. , Endele, S. , Schwarzmayr, T. , Albrecht, B. , Bartholdi, D. , Beygo, J. , Di Donato, N. , Dufke, A. , Cremer, K. , Hempel, M. , Horn, D. , Hoyer, J. , Joset, P. , Röpke, A. , Moog, U. , Riess, A. , … Strom, T. M. (2012). Range of genetic mutations associated with severe non‐syndromic sporadic intellectual disability: An exome sequencing study. Lancet, 380, 1674–1682. [DOI] [PubMed] [Google Scholar]
- Richards, S. , Aziz, N. , Bale, S. , Bick, D. , Das, S. , Gastier‐Foster, J. , Grody, W. W. , Hegde, M. , Lyon, E. , Spector, E. , Voelkerding, K. , Rehm, H. L. , & ACMG Laboratory Quality Assurance, C . (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs, E. R. , Andersen, E. F. , Cherry, A. M. , Kantarci, S. , Kearney, H. , Patel, A. , Raca, G. , Ritter, D. I. , South, S. T. , Thorland, E. C. , Pineda‐Alvarez, D. , Aradhya, S. , & Martin, C. L. (2020). Technical standards for the interpretation and reporting of constitutional copy‐number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genetics in Medicine, 22, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P. N. , Ravanmehr, V. , Jacobsen, J. O. B. , Danis, D. , Zhang, X. A. , Carmody, L. C. , Gargano, M. A. , Thaxton, C. L. , UNC Biocuration, C ., Karlebach, G. , Reese, J. , Holtgrewe, M. , Holtgrewe, M. , Köhler, S. , McMurry, J. A. , Haendel, M. A. , Smedley, D. (2020). Interpretable clinical genomics with a likelihood ratio paradigm. American Journal of Human Genetics, 107, 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, J. M. , Rödelsperger, C. , Schuelke, M. , & Seelow, D. (2010). MutationTaster evaluates disease‐causing potential of sequence alterations. Nature Methods, 7, 575–576. [DOI] [PubMed] [Google Scholar]
- Sifrim, A. , Popovic, D. , Tranchevent, L. C. , Ardeshirdavani, A. , Sakai, R. , Konings, P. , Vermeesch, J. R. , Aerts, J. , De Moor, B. , & Moreau, Y. (2013). eXtasy: Variant prioritization by genomic data fusion. Nature Methods, 10, 1083–1084. [DOI] [PubMed] [Google Scholar]
- Smedley, D. , Jacobsen, J. O. , Jäger, M. , Köhler, S. , Holtgrewe, M. , Schubach, M. , Siragusa, E. , Zemojtel, T. , Buske, O. J. , Washington, N. L. , Bone, W. P. , Haendel, M. A. , & Robinson, P. N. (2015). Next‐generation diagnostics and disease‐gene discovery with the Exomiser. Nature Protocols, 10, 2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley, D. , Schubach, M. , Jacobsen, J. O. B. , Köhler, S. , Zemojtel, T. , Spielmann, M. , Jäger, M. , Hochheiser, H. , Washington, N. L. , McMurry, J. A. , Haendel, M. A. , Mungall, C. J. , Lewis, S. E. , Groza, T. , Valentini, G. , & Robinson, P. N. (2016). A whole‐genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. American Journal of Human Genetics, 99, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley, D. , Smith, K. R. , Martin, A. , Thomas, E. A. , Mcdonagh, E. M. , Cipriani, V. , Ellingford, J. M. , Arno, G. , Tucci, A. , Vandrovcova, J. , Chan, G. , Williams, H. J. , Ratnaike, T. , Wei, W. , Stirrups, K. , Ibanez, K. , Moutsianas, L. , Wielscher, M. , … Gibson, K. (2021). The 100,000 genomes pilot on rare disease diagnosis in healthcare—A preliminary report. New England Journal of Medicine, 385, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter, K. , Adams, D. R. , Bacino, C. A. , Bellen, H. J. , Bernstein, J. A. , Cheatle‐Jarvela, A. M. , Eng, C. M. , Esteves, C. , Gahl, W. A. , Hamid, R. , Jacob, H. J. , Kikani, B. , Koeller, D. M. , Kohane, I. S. , Lee, B. H. , Loscalzo, J. , Luo, X. , McCray, A. T. , Metz, T. O. , … Undiagnosed Diseases, N . (2018). Effect of genetic diagnosis on patients with previously undiagnosed disease. New England Journal of Medicine, 379, 2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammimies, K. , Marshall, C. R. , Walker, S. , Kaur, G. , Thiruvahindrapuram, B. , Lionel, A. C. , Yuen, R. K. , Uddin, M. , Roberts, W. , Weksberg, R. , Woodbury‐Smith, M. , Zwaigenbaum, L. , Anagnostou, E. , Wang, Z. , Wei, J. , Howe, J. L. , Gazzellone, M. J. , Lau, L. , Sung, W. W. , … Fernandez, B. A. (2015). Molecular diagnostic yield of chromosomal microarray analysis and whole‐exome sequencing in children with autism spectrum disorder. Journal of the American Medical Association, 314, 895–903. [DOI] [PubMed] [Google Scholar]
- Thompson, R. , Papakonstantinou Ntalis, A. , Beltran, S. , Töpf, A. , de Paula Estephan, E. , Polavarapu, K. , 't Hoen, P. A. C. , Missier, P. , & Lochmüller, H. (2019). Increasing phenotypic annotation improves the diagnostic rate of exome sequencing in a rare neuromuscular disorder. Human Mutation, 40(10), 1797–1812. [DOI] [PubMed] [Google Scholar]
- Turnbull, C. , Scott, R. H. , Thomas, E. , Jones, L. , Murugaesu, N. , Pretty, F. B. , Halai, D. , Baple, E. , Craig, C. , Hamblin, A. , Henderson, S. , Patch, C. , O'Neill, A. , Devereau, A. , Smith, K. , Martin, A. R. , Sosinsky, A. , McDonagh, E. M. , Sultana, R. , … Genomes, P. (2018). The 100 000 Genomes Project: Bringing whole genome sequencing to the NHS. BMJ, 361, k1687. [DOI] [PubMed] [Google Scholar]
- Wu, C. , Devkota, B. , Evans, P. , Zhao, X. , Baker, S. W. , Niazi, R. , Cao, K. , Gonzalez, M. A. , Jayaraman, P. , Conlin, L. K. , Krock, B. L. , Deardorff, M. A. , Spinner, N. B. , Krantz, I. D. , Santani, A. B. , Tayoun, A. , & Sarmady, M. (2019). Rapid and accurate interpretation of clinical exomes using Phenoxome: A computational phenotype‐driven approach. European Journal of Human Genetics, 27, 612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , & Wang, K. (2015). Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nature Protocols, 10, 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Muzny, D. M. , Reid, J. G. , Bainbridge, M. N. , Willis, A. , Ward, P. A. , Braxton, A. , Beuten, J. , Xia, F. , Niu, Z. , Hardison, M. , Person, R. , Bekheirnia, M. R. , Leduc, M. S. , Kirby, A. , Pham, P. , Scull, J. , Wang, M. , Ding, Y. , … Eng, C. M. (2013). Clinical whole‐exome sequencing for the diagnosis of Mendelian disorders. New England Journal of Medicine, 369, 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Muzny, D. M. , Xia, F. , Niu, Z. , Person, R. , Ding, Y. , Ward, P. , Braxton, A. , Wang, M. , Buhay, C. , Veeraraghavan, N. , Hawes, A. , Chiang, T. , Leduc, M. , Beuten, J. , Zhang, J. , He, W. , Scull, J. , Willis, A. , … Eng, C. M. (2014). Molecular findings among patients referred for clinical whole‐exome sequencing. Journal of the American Medical Association, 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel, T. , Köhler, S. , Mackenroth, L. , Jäger, M. , Hecht, J. , Krawitz, P. , Graul‐Neumann, L. , Doelken, S. , Ehmke, N. , Spielmann, M. , Oien, N. C. , Schweiger, M. R. , Krüger, U. , Frommer, G. , Fischer, B. , Kornak, U. , Flöttmann, R. , Ardeshirdavani, A. , Moreau, Y. , … Robinson, P. N. (2014). Effective diagnosis of genetic disease by computational phenotype analysis of the disease‐associated genome. Science Translational Medicine, 6, 252ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Havrilla, J. M. , Fang, L. , Chen, Y. , Peng, J. , Liu, C. , Wu, C. , Sarmady, M. , Botas, P. , Isla, J. , Lyon, G. J. , Weng, C. , & Wang, K. (2020). Phen2Gene: Rapid phenotype‐driven gene prioritization for rare diseases. NAR Genomics and Bioinformatics, 2, lqaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Petrovski, S. , Xie, P. , Ruzzo, E. K. , Lu, Y.‐F. , McSweeney, K. M. , Ben‐Zeev, B. , Nissenkorn, A. , Anikster, Y. , Oz‐Levi, D. , Dhindsa, R. S. , Hitomi, Y. , Schoch, K. , Spillmann, R. C. , Heimer, G. , Marek‐Yagel, D. , Tzadok, M. , Han, Y. , Worley, G. , … Goldstein, D. B. (2015). Whole‐exome sequencing in undiagnosed genetic diseases: Interpreting 119 trios. Genetics in Medicine, 17, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described in the paper are already provided in the paper except for access to the 100,000 Genomes Project samples which is by application to Genomics England.


