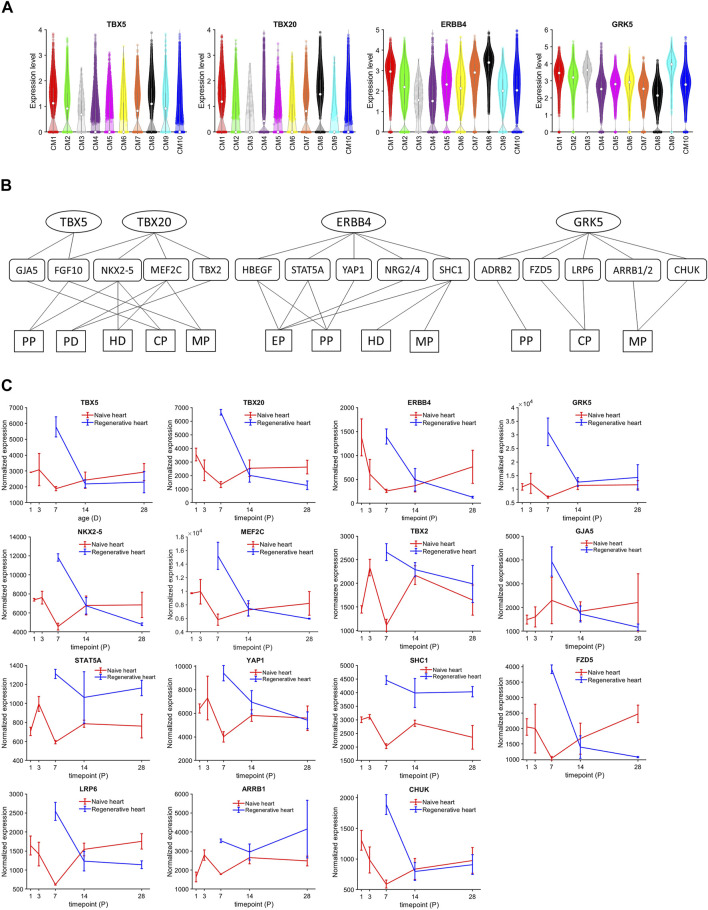
FIGURE 3.
TBX5, TBX20, ERBB4, and GRK5 were consistently upregulated in CM1, CM7, and CM8 cardiomyocytes. (A) The abundance of TBX5, TBX20, ERBB4, and GRK5 RNA transcripts was measured in cardiomyocytes from each cluster and displayed in a violin plot. The expression was normalized according to the Seurat pipeline: the expression matrix was scaled by the ScaleData function with vars. to. regress set to nUMI and nGenes; then, the scaled expression was log-normalized. (B) The network of genes, cellular processes, and signaling pathways regulated by TBX5, TBX20, ERBB4, and GRK5 was evaluated with DAVID bioinformatics resources. Targeted genes were identified in the TRRUST v2 (TBX5 and TBX20) and STRING v11.5 (ERBB4 and GRK5) databases and functional/pathway data were obtained from the GOA, KEGG, and Reactome databases. PP, positive regulation of cell proliferation; PD, positive regulation of cardiac muscle cell differentiation; CP, canonical Wnt signaling pathway; MP, MAPK signaling pathway; Ep, ErbB signaling pathway. (C) The bulk RNAseq expressions of TBX5, TBX20, ERBB4, GRK5, NKX2-5, MEF2C, TBX2, GJA5, STAT5A, YAP1, SHC1, FZD5, LRP5, ARRB1, and CHUK in regenerative and naïve pig heart. The transcript counts were normalized using DeSeq2 pipeline.