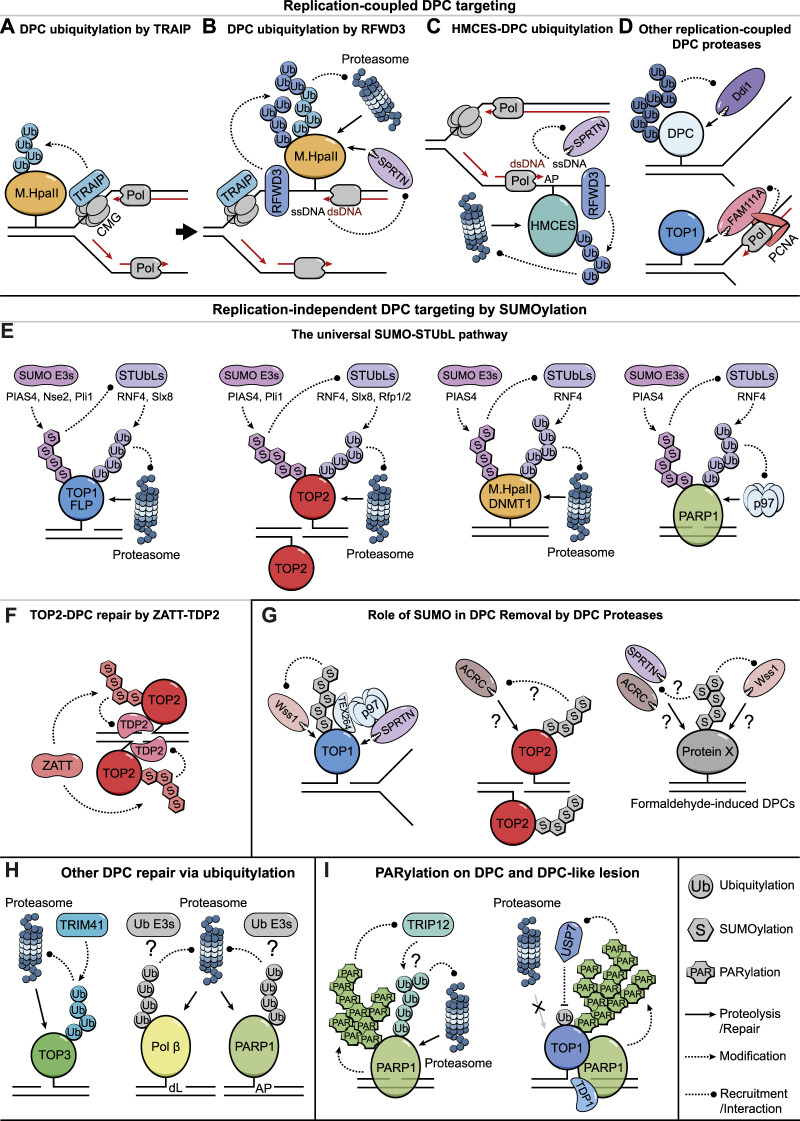
FIGURE 1.
Targeting DPCs for Removal via PTMs. (A) A schematic illustration of TRAIP-mediated ubiquitylation of DPCs that hinder CMG progression. (B) CMG bypass of DPCs exposes ssDNA and likely triggers RFWD3-mediated DPC ubiquitylation that further leads to proteolysis by the proteasome and SPRTN. SPRTN protease is targeted by ssDNA/dsDNA junctions. (C) Putative models illustrating how HMCES-DPCs on AP sites are either ubiquitylated by RFWD3 to undergo proteasomal degradation or targeted by SPRTN via nascent DNA strands extended to the lesion. (D) Ddi1/DDI2 might target DPCs with long ubiquitin chains, presumably associated with DNA replication (top illustration). FAM111A degrades DPCs during DNA replication via its interaction with PCNA (bottom illustration). (E) The SUMO-STUbL pathway targets DPCs to degradation in the absence of DNA replication and serves as a universal repair solution for various types of DPCs and DPC-like lesions. (F) ZATT-mediated TOP2-DPC SUMOylation facilitates the recruitment of TDP2 to hydrolyze the covalent linkages. (G) Role of SUMOylation in promoting the removal of TOP1/2-DPCs and formaldehyde-induced DPCs by DPC proteases. (H) Ubiquitylation-mediated proteasomal degradation of crosslinked TOP3B, Polβ-, and PARP1-DPCs. (I) PARP1 auto-PARylation limits PARP1-traping via TRIP12-mediated ubiquitylation and proteasomal degradation (left illustration). PARylation stimulates TDP1 recruitment to TOP1-DPCs while also preventing their proteasomal degradation by recruiting the deubiquitylating enzyme USP7 (right illustration).