Summary
Over a time period of 18 months an Enterobacter hormaechei sequence type (ST) 90, harboring a blaOXA-436 carbapenemase gene, was isolated from seven patients at Odense University Hospital, Denmark. The patients were all from the same department, but there was no apparent direct epidemiological link. Whole genome sequencing (WGS) was performed on all clinical isolates as well as on a number of environmental samples including two E. hormaechei ST90 isolates carrying the blaOXA-436 gene, which were isolated in samples from two shower drains at the department. These drains were suspected to be the source of the outbreak.
Keywords: Carbapenemase producing organisms, Enterobacter hormaechei, Environmental transmission, Whole genome sequencing
Introduction
The incidence of clinical infections due to carbapenemase producing organisms (CPO) is still low in Denmark and often related to a history of international travel [1]. However, the incidence has increased in the past few years and in 2020, 238 CPOs were detected from 207 patients with the majority of these being carbapenemase producing enterobacterales (CPE) [1]. Currently all carbapenemase producing isolates are submitted for verification and genotyping by WGS at the National Reference Laboratory for Antimicrobial Resistance at Statens Serum Institut, Copenhagen, Denmark. Carbapenemase producing organisms are (as of September 2018) notifiable according to the Danish Health Authorities. Larger outbreaks of CPEs have been described within and between hospitals and regions in Denmark [1]. Patients are not routinely screened for CPOs in Denmark unless the patient has a recent travel history outside the Nordic countries.
In August 2017 an Enterobacter hormaechei sequence type (ST) 90 (E. cloacae complex MLST scheme) harboring a blaOXA-436 (blaOXA-48 like) carbapenemase gene was isolated from a wound swab, of a patient (index), who had recently been discharged from the Department of Cardiology, Odense University Hospital, Denmark [2]. The gene was located on a large IncHI1/IncHI2 plasmid and this specific combination of sequence type and gene/plasmid had not been observed in Denmark previously. However, the associated plasmid (pOXA436) had been described in E. asburiae from Denmark in 2013 as well as Citrobacter freundii and Klebsiella pneumoniae in 2014 from a total of four patients [3]. In this current case the patient had no reported history of recent travel outside Denmark or CPE carriage prior to the ST90 E. hormaechei isolate. During the following 18 months, six other patients were diagnosed with the same E. hormaechei ST90 with the blaOXA-436 gene (three from blood cultures, one swab from an IV-catheter puncture site and two urine samples). Apart from the two first patients (hospitalized at the same time), there was no direct epidemiological link between the patients, but the common denominator was hospitalization in the same department of cardiology (Figure 1).
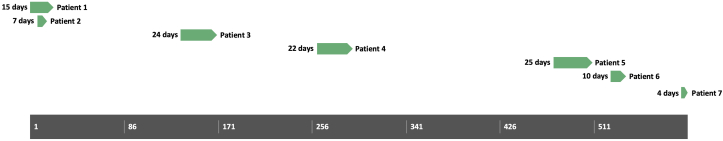
Figure 1.
Timeline picturing the duration of the outbreak in total number of days. Green bars show the total days of admittance at the department of cardiology for each patient. Except for the first two patients, there is no overlap of hospitalization between the patients.
In the 18 month period, hand hygiene audits were performed, usual hygiene measures were reviewed by the members of the infection control team at the hospital and all admitted patients were screened with rectal swabs several times during the outbreak. Furthermore, after the third patient was identified in January 2018, a thorough cleaning and disinfection of selected rooms with vaporized hydrogen peroxide (RHEA Compact, airinspace, Élancourt, France) was performed.
After patients number six and seven were identified in February 2019 an environmental reservoir of the organism was suspected. This resulted in further measures, including replacement of curtains between patient beds with washable screens, thorough cleaning of all equipment, storage of equipment in closed closets and the introduction of single use equipment when possible. Personnel in the department were not screened at any point during the outbreak. Finally, it was also decided to collect environmental samples from the department.
Methods
The environmental screening included two samples from shower drains, five samples from floor drains below sinks, 25 samples from sinks and three samples from bedpan boilers/instrument washers. All 35 sites were swabbed (eSwab, Copan, Brescia, Italy). The swabs were inserted as deep as possible into the sample object and rotated 20–30 times. All swabs were vortexed and cultured directly from the eSwab onto CHROMID Carba Smart selective chromogenic media bi-plate (bioMerieux, Craponne, France), with 100 μl eSwab media on each side of the two-sided plate, and incubated for 24 hours. Colonies were identified using MALDI-TOF (Bruke Daltonik, Bremen Germany). All isolates belonging to the family Enterobacteriales were further screened with the rapid test NG Test CARBA 5 (NG Biotech, Guipry, France), which can detect the most common carbapenemases (including the OXA-48 like enzymes such as OXA-436) within 15 minutes. Susceptibility testing with disk diffusion according to EUCAST was performed on all NG Test CARBA 5 positive isolates to confirm carbapenemase production and the isolates were sent for WGS at the Danish National Reference Laboratory for Antimicrobial Resistance at Statens Serum Institut (Copenhagen). Here, genomic DNA was extracted from the isolates (DNeasy Blood and Tissue Kit, Qiagen, Copenhagen, Denmark), following construction of fragment libraries by using the Nextera XT Kit (Illumina, Little Chesterford, UK). Paired-end sequencing with read lengths of 251 bp was performed (MiSeq, Illumina) according to the manufacturer's instructions. Draft assembly and detection of antimicrobial resistance genes were performed using the BIFROST QC and analysis pipeline (“https://github.com/ssi-dk/bifrost”). For Nanopore sequencing, DNA was extracted with the GenFind V2 kit (Beckman Coulter) using a DynaMag-2 magnet (Thermo Fisher Scientific). A library was prepared using the protocol ‘1D Native barcoding genomic DNA (with #EXP-NBD103 and #SQK-LSK109)’ (Oxford Nanopore Technologies, United Kingdom) and sequenced in a R9.41 flow cell (FLO-MIN106-D) inside a MinION Mk1B. Using Guppy v4.2.2 (ONT) raw fast5 reads were base-called to fastq format in ‘high-accuracy methylation-aware’ configuration, demultiplexed and quality filtered to minimum q10. Then illumina-nanopore hybrid de novo genome assembly was run with Unicycler v0.4.8-beta [4].
The raw short-read sequencing data of the two OXA-436-producing ST90 E. hormaechei isolated from shower drains in this study were uploaded to the CSIphylogeni 1.4 website [5]. This was done together with the seven patient OXA-436-producing ST90 E. hormaechei. Four additional (non-related and originating from different Danish hospitals) ST90 E. hormaechei out-group isolates were tested. These isolates were carrying either a blaVIM-1 carbapenemase or no carbapenemase gene for SNP-based cluster analysis using the draft genome of the drain isolate (ST90_D1) as reference.
Results
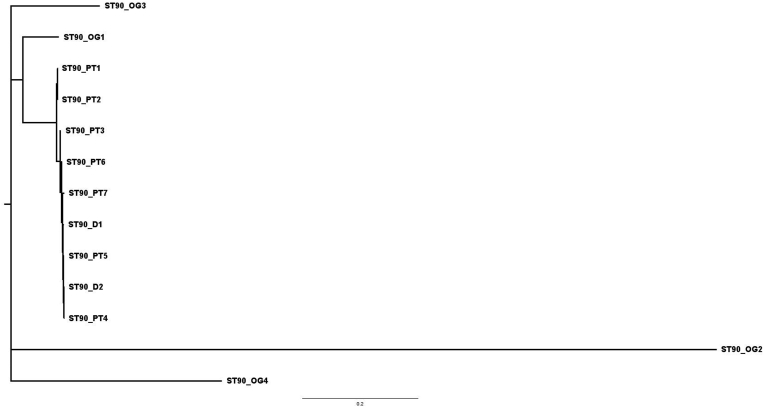
Two E. hormaechei ST90 isolates (hereafter named ST90 D1 and ST90 D2) carrying the blaOXA-436 gene were isolated in the environmental samples taken from the shower drains in the only two patient bathrooms in the unit. The two bathrooms were adjacent to each other. Phylogenetic analysis using whole genome sequencing short-read data of the two drain isolates together with sequencing data originating from a total of seven patient isolates (named ST90 PT1 to ST90 PT7) of the same MLST type and also carrying blaOXA-436 as well as sequencing data from four non-related ST90 E. hormaechei out-group isolates (named ST90 OG1 to ST90 OG4) was performed. The analysis showed that the two drain isolates were closely related (between 0 and 11 SNPs) to the seven blaOXA-436 -positive isolates but unrelated to the four isolates carrying a different carbapenemase (113–569 SNPs covering 89.4% of the reference) (Figure 2). Through long-read sequencing on the ONT MinION platform, the complete sequence of the plasmid carrying the blaOXA-436 gene was obtained from one of the drain isolates (ST90 D1) and compared to the previously sequenced pOXA-436 plasmid (GenBank accession number KY863418) originating from an E. aspuriae isolated in 2013 from a patient in the Capital region of Denmark [3]. The plasmid from the drain isolate had a 99.7% sequence identity to pOXA436 and only deviated from this plasmid by acquisition of a single IS3 element and loss of another element (IS3) (Figure 2).
Figure 2.
Maximum likelihood tree created in FigTree and based on SNP distances of whole genome sequencing of the seven isolates from patients (PT1-PT7), the two drain isolates (D1-D2) and four non-related out-group ST90 isolates (OG1-OG4).
Furthermore two Citrobacter freundii (ST534), with the same blaOXA-436 carbapenemase gene, were detected from two sinks in a wash room in the department. The wash room is used for emptying and cleaning bedpans. Citrobacter freundii ST534 has not been observed in the national surveillance of CPEs in Denmark, but subsequent long-read sequencing revealed that the blaOXA-436 gene of the C. freundii ST534 strain were also located on a large plasmid highly similar to pOXA-436 (data not shown).
Discussion
After the recovery of the outbreak strain from the shower drains, staff reported that these drains had a tendency to become partly blocked resulting in regular overflow of water from the drains while patients were showering. The drains therefore seemed to be a potential source of this outbreak as no existing interventions had curtailed the outbreak. The hospitalestates department was involved and the floor grate and traps were changed and fixed to the drain, so that they could not be removed and contaminate other rooms (discussed below). The bathrooms were emptied and cleaned. The part of the floor drains, that wasn't possible to change were manually cleaned and afterward rinsed with vinegar. Finally the bathrooms were disinfected with vaporized hydrogen peroxide (RHEA Compact) following cleaning. The shower heads were relocated so the patients didn't have to stand on top of the drain while showering and the water jet didn't hit the drain directly; thereby reducing the water splash. The technical department cleaned the waste pipes and reestablished the function of the drains and sewer system to prevent overflow. In addition to the regular cleaning of the two bathrooms, an extra daily cleaning with chlorine disinfection of all contact points was established.
Prior to this outbreak, the traps from the bathrooms had occasionally been cleaned in the wash room sinks (where the isolates of the C. freundii carrying the same blaOXA-436 carbapenemase gene had been detected). This might indicate that a plasmid with the blaOXA-436 gene was transferred from the outbreak E. hormaechei ST90 to the C. freundii either in the sink or in the wash room traps or drains (or vice versa). The E. hormaechei itself was never recovered from these two wash room sinks. The C. freundii sink strain has never been detected in any samples from patients, neither at Odense University Hospital nor in Denmark. Cleaning traps in any sink generally risks thespread of bacteria from one part of a department to another and thereby increases the risk of in-ward transmission.
In June and September 2020, almost three years after the first discovery of E. hormaechei blaOXA-436, new samples were taken from the drains in the two bathrooms and plated on CHROMID Carba Smart. No E. hormaechei or other Enterobacteriales were recovered from these samples and no further patients have been diagnosed with E. hormaechei blaOXA-436 at Odense University Hospital since March 2019.
The incidence of CPO is still low in Denmark and often associated with travelling. Since CPOs have been made notifiable, and all Danish CPO isolates are sent to the reference laboratory at Statens Serum Institut, it is possible to survey the incidence and outbreaks on a national level. Here, whole genome sequencing is a powerful tool to determine whether an incident is part of an outbreak.
This report shows that outbreaks can last over a long period of time and can be difficult to control and resolve. In this case, a likely source of the outbreak was the two shower drains as the outbreak was terminated after the drains had been replaced. Other explanations may well exist, but It is well known, that the hospital environment and sewer systems can be a source of transmission [[6], [7], [8], [9], [10]]. This outbreak underlines the importance of maintaining department facilities and sewer systems and that staff are trained to report non-functioning installations, in addition to general hygiene measures to prevent transmission of bacteria between patients. In Denmark, outbreaks or incidents of CPO with no prior history of travelling should make one aware of the risk of in-ward transmission.
Credit author statement
All: Writing - Review & Editing.
Christina Raun-Petersen: Conceptualization, Investigation, writing – original draft.
Anette Toft: Methodology, Investigation.
Mette Marie Nordestgaard: Methodology.
Anette Holm: Methodology, Investigation.
Søren Overballe-Petersen: Formal analyses.
Anette Hammerum: Formal analyses.
Henrik Hasman: Formal analyses.
Ulrik Stenz Justesen: Conceptualization, Methodology, Investigation, writing – original draft, Supervision.
Conflict of interest statement
None declared.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.DANMAP. DANMAP 2020 – Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans I Denmark. https://www.danmap.org/reports/2020 [accessed October 12, 2021].
- 2.Miyoshi-Akiyama T., Hayakawa K., Ohmagari N., Shimojima M., Kirikae T. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuelsen Ø., Hansen F., Aasnæs B., Hasman H., Lund B.A., Leiros H.S., et al. Dissemination and Characteristics of a Novel Plasmid-Encoded Carbapenem-Hydrolyzing Class D β-Lactamase, OXA-436, Found in Isolates from Four Patients at Six Different Hospitals in Denmark. Antimicrob Agents Chemother. 2017;62(1) doi: 10.1128/AAC.01260-17. e01260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6) doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chia P.Y., Sengupta S., Kukreja A., Ponnampalavanar S.S.L., Ng O.T., Marimuthu K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob Resist Infect Control. 2020;9(1):29. doi: 10.1186/s13756-020-0685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Man T.J.B., Yaffee A.Q., Zhu W., Batra D., Alyanak E., Rowe L.A., et al. Multispecies Outbreak of Verona Integron-Encoded Metallo-β-Lactamase-Producing Multidrug Resistant Bacteria Driven by a Promiscuous Incompatibility Group A/C2 Plasmid. Clin Infect Dis. 2021;72(3):414–420. doi: 10.1093/cid/ciaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemarié C., Legeay C., Mahieu R., Moal F., Ramont C., Kouatchet A., et al. Long-term contamination of sink drains by carbapenemase-producing Enterobacterales in three intensive care units: characteristics and transmission to patients. J Hosp Infect. 2021;112:16–20. doi: 10.1016/j.jhin.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Heireman L., Hamerlinck H., Vandendriessche S., Boelens J., Coorevits L., Brabandere E.D., et al. Toilet drain water as a potential source of hospital room-to-room transmission of carbapenemase-producin klebsiella pneumoniae. J Hosp Infect. 2020;106(2):232–239. doi: 10.1016/j.jhin.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Jolivet S., Couturier J., Vuillemin X., Gouot C., Nesa D., Adam M., et al. Outbreak of OXA-48-producing Enterobacterales in a haematological ward associated with an uncommon environmental reservoir, France, 2016 to 2019. Euro Surveill. 2021;26(21) doi: 10.2807/1560-7917.ES.2021.26.21.2000118. [DOI] [PMC free article] [PubMed] [Google Scholar]