Abstract
The whole world is still challenged with COVID-19 pandemic caused by Coronavirus-2 (SARS-CoV-2) which has affected millions of individuals around the globe. Although there are prophylactic vaccines being used, till now, there is ongoing research into discovery of drug candidates for total eradication of all types of coronaviruses. In this context, this study sought to investigate the inhibitory effects of six selected tropical plants against four pathogenic proteins of Coronavirus-2. The medicinal plants used in this study were selected based on their traditional applications in herbal medicine to treat COVID-19 and related symptoms. The biological activities (antioxidant, free radical scavenging, and anti-inflammatory activities) of the extracts of the plants were assessed using different standard procedures. The phytochemicals present in the extracts were identified using GCMS and further screened via in silico molecular docking. The data from this study demonstrated that the phytochemicals of the selected tropical medicinal plants displayed substantial binding affinity to the binding pockets of the four main pathogenic proteins of Coronavirus-2 indicating them as putative inhibitors of Coronavirus-2 and as potential anti-coronavirus drug candidates. The reaction between these phytocompounds and proteins of Coronavirus-2 could alter the pathophysiology of COVID-19, thus mitigating its pathogenic reactions/activities. In conclusion, phytocompounds of these plants exhibited promising binding efficiency with target proteins of SARS-COV-2. Nevertheless, in vitro and in vivo studies are important to potentiate these findings. Other drug techniques or models are vital to elucidate their compatibility and usage as adjuvants in vaccine development against the highly contagious COVID-19 infection.
Keywords: COVID-19, Coronavirus-2, Medicinal plants, Phytochemicals, In silico molecular docking
Introduction
For several decades, infectious diseases particularly those induced by pathogenic viruses have a significant devastating effect on human wellbeing and health (Oladele et al. 2020a, b, c, d). At present, the whole world is challenged with COVID-19 pandemic caused by Coronavirus-2 (SARS-CoV-2) which has affected millions of individuals around the globe (Huang et al. 2020; Oladele et al. 2021a, b). The SARS-CoV-2 is a betacoronavirus and a member of the Coronaviridae family (Wang et al. 2020). Although there are prophylactic vaccines being used, till now, there is ongoing research into drug candidate for total eradication of all types of coronaviruses and therapy or medication to inhibit the progression of the pathogenesis of the disease. Thus, there is urgent need for the discovery of non-invasive, novel, natural, and non-toxic therapeutic agents that will effectively suppress the pathogenesis of Coronavirus-2.
SARS-CoV-2 belongs to either the alpha or beta human coronaviruses; several processes including pathogenesis, infectivity, virulence, and virus release are mediated by unique structural and accessory proteins (Issa et al 2020; Hassan et al. 2021). Worthy of note are the main protease, primase, ADP ribose phosphatase, replicase protein, and RNA-dependent RNA polymerase (Das et al. 2021; Maio et al. 2021). The activity of the main protease (Mpro), a 33.8 kDa enzyme is highly integral to the release of the viruses’ functional polypeptide via proteolytic processing necessary for its replication and transcription (Zhou et al. 2020), unlike the ADP ribose phosphatase and replicase protein, which are associated with altering the immune response of the host by removal of ADP-ribose from ADP-ribosylated proteins and viral replication (Michalska et al. 2020). Functionally similar to Mpro, the RNA-dependent RNA polymerase has been implicated in the replication of virus genome and gene transcription, making these proteins a potential candidate drug targets in mitigating viral lethality (Maio et al. 2021).
Medicinal plants are used since time immemorial in the treatment of various diseases due to their accessibility, inexpensive natural products, and host broad range of both biologically and chemically active phytocompounds which have been used in the synthesis of therapeutic drugs (Oladele et al. 2020a,b, 2021a, b). They also serve as sources of adjuvants for vaccine development. The six medicinal plants (Neem [Azadirachta indica Juss], Jute [Corchorus olitorius], Cassia alata, Phyllanthus amarus, bitter leaf [Vernonia amygdalina], and Cashew [Anacardium occidentale L.]) used in this study have been reported to have antioxidant, antiviral, immunomodulatory, cytoprotective, and antipyretic properties. Mode of actions of SARS-CoV-2 includes immunosuppression, alteration to physiological functions of alveoli, and activation of c-Jun N-terminal kinase (JNK) and mitogen-activated protein kinase (MAPK) pathways leading to secondary hemophagocytic lymphohistiocytosis (sHLH), a cytokine profile with a hyperinflammatory syndrome associated with multiorgan failure linked to COVID-19 severity including lung damage (Oladele et al. 2020c).
Based on the aforementioned, this study sought to investigate the cytoprotective effects of six tropical medicinal plants used in traditional medicine to treat COVID-19-related symptoms and examine the inhibitory ability of the active natural products and phytochemicals present in these plants against five essential proteins implicated in the pathogenesis of COVID-19. Furthermore, the study explores the in vitro biological properties of the plants aqueous extracts.
Materials and methods
Materials
Bovine serum albumin (BSA), folin ciocalteau, Tris–HCl, ethylene diaminetetraacetic acid (EDTA), phenyl methyl sulfonyl fluoride (PMSF), DPPH (1,1-diphenyl-1,2-picrylhydrazyl), TPTZ (2,4,6,-tripyridyl-s-triazine), and deoxyribose were purchased from Sigma Chemical Company, St Louis, Mo, USA. Hydrochloric acid (HCl), methanol, gallic acid, sulphuric acid (H2SO4), sodium carbonate (Na2CO3), aluminium chloride, potassium acetate, potassium persulphate, sodium nitroprusside, hydrogen peroxide, glacial acetic acid, naphthylethylenediamine dichloride, NADH, trichloroacetic acid (TCA), thiobarbituric acid (TBA), and L-ascorbic acid were all purchased from Merck, USA. All other chemicals and reagents used were of analytical grade.
Extraction of the aqueous extracts of the plants
The six tropical medicinal plants (Azadirachta indica Juss, Corchorus olitorius, Cassia alata, Phyllanthus amarus, Vernonia amygdalina, and Anacardium occidentale L.) used in this study are currently used in traditional and herbal medicine to treat and/or prevent COVID-19-related symptoms (Oladele et al. 2020a, b, c, d). Fresh leaves of Azadirachta indica, Corchorus olitorius, Cassia alata, Phyllanthus amarus, Vernonia amygdalina, and bark of Anacardium occidentale were air dried at room temperature in the Biochemistry laboratory of the Department of Chemical Sciences, Faculty of Science, Kings University, Odeomu, Osun State. The dried leaves were pulverized using electric blender. The powdered sample was weighed and soaked in 6 volumes of distilled water for 72 h with constant stirring. The extract was obtained by filtration and the paste was then freeze dried at − 60 °C.
Assessment of biological properties of the aqueous extracts of the plants
Nitric oxide radical scavenging activity
The scavenging effect of aqueous extracts of each plant on nitric oxide radical was measured according to the method of Ebrahimzadeh et al. (2009). Aliquot (1 mL) of sodium nitroprusside (5 mM) in phosphate-buffered saline (PBS, pH 7.2) was mixed with different concentrations of each plant aqueous extracts. This was incubated at room temperature for 150 min after which 0.5 mL of Griess reagent was added. The absorbance of the pink chromophore formed was read at 546 nm. Gallic acid and ascorbic acid were used as positive control. All experiment was done in triplicates. The percentage inhibition was calculated as follows:
Lipid peroxidation inhibition assay
The lipid peroxidation inhibition assay (LPI) was determined according to the method described by Liu et al. (2003) with a slight modification. Excised rat liver was homogenized in phosphate buffer (pH 7.4) and then centrifuged to obtain liposome. Aliquot (0.5 mL) of supernatant, 100 μL 10 mM FeSO4, 100 μL 0.1 mM ascorbic acid, and 0.3 mL of each plant aqueous extracts or standard at different concentrations were mixed to make the final volume of 1 ml. The reaction mixture was incubated at 37 °C for 20 min. Aliquot (1 mL) of (2%) TCA and 1.5 mL of (1%) TBA were added immediately after heating. Finally, the reaction mixture was heated at 100 °C for 15 min and cooled to room temperature. The absorbance was then taken at 532 nm. Percentage inhibition of lipid peroxidation (% LPI) was calculated as follows:
where Acontrol is the absorbance of the control and Asample is the absorbance of the extractive/standard. Percentage of inhibition was plotted against concentration.
Ferric reducing power assay
Slightly modified assay method described by Shahi et al. (2020) was employed. Typically, aliquots of the aqueous extracts of each plant were mixed with 0.2 mM phosphate buffer (0.5 mL; pH 6.6) and 0.5 mL potassium ferricyanide (1% w/v). The mixture was incubated at 50 °C for 20 min. Trichloroacetic acid (10%, 2.5 mL) was added to the mixture and centrifuged at 2500 rpm for 10 min. Aliquot (1 mL) of obtained supernatant was mixed with 1 mL FeCl3 (0.1%, 0.5 mL), and the absorbance was measured at 700 nm. Gallic acid and ascorbic acid were used as positive standard.
Hydroxyl radical scavenging activity
The hydroxyl radical scavenging activity of aqueous extracts of each plant was carried out using the assay method of Kim and Minamikawa (1997) as described by Shahi et al. (2020) with slight modifications. Briefly, 0.1 mL of 10 mM EDTA, 0.01 mL of 10 mM FeCl3, 0.1 mL of 10 mM H2O2, 0.36 mL of 10 mM deoxyribose, 1.0 mL of aqueous extracts of each plant, 0.33 mL of phosphate buffer (50 mM, pH 7.4), and 0.1 mL of ascorbic acid were added sequentially. Resulting mixture was then incubated at 37 °C for 1 h. Aliquot (1.0 mL) of 5% TCA and 1.0 mL of 0.5% TBA were added to develop the pink chromogen measured at 532 nm. The standard was gallic acid and ascorbic acid. The percentage of hydroxyl radical scavenged was estimated using the following equation:
where Acontrol is the absorbance of control and Asample is the absorbance of sample solution.
DPPH radical scavenging assay
Assay method of Wu et al. (2003) with slight modifications was employed. Briefly, aliquots (1 mL) of varying concentrations (50–800 mg/mL) of aqueous extracts of each plant were added to 1 mM 2, 2 diphenyl-1-picryldydrazyl (DPPH) (1 mL) in methanol solution. Resulting mixtures were vortexed, centrifuged (2500 rpm for 10 min), and then incubated in a dark chamber for 30 min at room temperature. Absorbance of the supernatant was measured at 517 nm against DPPH control containing 1 mL methanol in place of the protein fractions. Gallic acid and ascorbic acid were used as positive standard. Inhibition of DPPH radical was calculated as a percentage using the expression:
where Ablank was standard absorbance and Asample was sample absorbance.
Scavenging of hydrogen peroxide
The ability of the aqueous extracts of each plant to scavenge hydrogen peroxide was determined according to Nabavi et al. (2008) and (2009). A solution of hydrogen peroxide (40 mM) was prepared in phosphate buffer (pH 7.4). The concentration of hydrogen peroxide was determined by absorption at 230 nm using a spectrophotometer. Aqueous extracts of each plant (50–800 mg/ ml) in distilled water were added to a hydrogen peroxide solution (0.6 mL, 40 mM). The absorbance of hydrogen peroxide at 230 nm was determined after 10 min against a blank solution containing phosphate buffer without hydrogen peroxide. The percentage of hydrogen peroxide scavenging by the aqueous extracts of each plant and a standard compound was calculated as follows:
where Ao was the absorbance of the control and A1 was the absorbance in the presence of the sample of aqueous extracts of each plant and standard.
Gas chromatography-mass spectroscopy (GC–MS) analysis of the extract
The GC–MS analysis was carried out to determine the phytochemicals present in the aqueous extracts using GC–MS (Model: QP2010 plus Shimadzu, Japan) encompassing an AOC-20i auto-sampler and gas chromatograph interfaced to a mass spectrometer (GC–MS). The relative percentage of the analyte was expressed as a percentage with peak area normalization. Interpretation on the mass spectrum was conducted using the database of National Institute of Standards and Technology (NIST). The fragmentation pattern spectra of the unknown components were compared with those of known components stored in the NIST library (NIST 11). The relative percentage of each phytochemical was calculated by comparing its average peak area to the total area. The name, molecular weight, and structure of the components of the test materials were ascertained.
In silico studies
Protein preparation
The crystal structures of SARS coronavirus spike receptor-binding domain (PDB ID 2AJF) with resolution 2.90 Å, ADP ribose phosphatase of NSP3 from SARS CoV-2 (PDB ID 6VXS) with resolution 2.03 Å, SARS-COV2 major protease (PDB ID 6LU7) with resolution 2.16 Å, and SARS CoV-2 RNA-dependent RNA polymerase (PDB ID 7BTF) with resolution 2.95 Å were retrieved from the protein databank (www.rcsb.org). Before the docking and analysis, the crystal structures were processed by eliminating existing ligands and water molecules. Furthermore, missing hydrogen atoms were added using Autodock v4.2 program, Scripps Research Institute (Morris et al. 2009). Subsequently, non-polar hydrogens were merged while polar hydrogen was added and then saved into pdbqt format in preparation for molecular docking.
Ligand preparation
The SDF structures of phytochemical present in the extracts as identified by GC–MS were retrieved from the PubChem database (www.pubchem.ncbi.nlm.nih.gov). The phytochemicals were converted to mol2 chemical format. Polar hydrogens were added while non-polar hydrogens were merged with the carbons, and the internal degrees of freedom and torsions were set. The protein and ligand molecules were further converted to the dockable PDBQT format using Autodock tools.
Molecular docking
Docking of the phytochemicals to the targeted protein and determination of binding affinities was carried out using AutodockVina (Trott and Olson 2010). The PDBQT formats of the receptor and that of the phytochemicals were positioned at their respective columns, and the software was run. The binding affinities of phytochemicals for the protein target were recorded. The phytochemicals were then ranked by their affinity scores. The molecular interactions between the receptor and phytochemicals with most remarkable binding affinities were viewed with PYMOL.
ADMET analysis
The solubility, pharmacodynamics, pharmacokinetics, and toxicological profiles of phytochemicals with the best docking score were computed based on their ADMET (absorption, distribution, metabolism, elimination, and toxicity) studies using pkCSM tool (http://biosig.unimelb.edu.au/pkcsm/prediction). The canonical SMILE molecular structures of the compounds used in the studies were obtained from PubChem (pubchem.ncbi.nlm.nih.gov).
Results
Biological properties of the aqueous extracts of the plants
Nitric oxide inhibitory activity of the aqueous extracts of the plants
Figure 1 shows a significant increase in the nitric oxide inhibitory activity of all aqueous extracts of the selected medicinal plants at various concentrations used in this study. At 800 mg/ml, ALVA (aqueous leaf extract of Vernonia amygdalina) displayed the highest nitric oxide inhibitory activity among the six plant extracts. Also, ALVA exhibited higher activity than ascorbic acid but not up to activity of the gallic acid. This confirms the possible anti-inflammatory potential of the aqueous extracts.
Fig. 1.
Nitric oxide inhibitory activity of aqueous extracts of selected medicinal plants. ALAI, aqueous leaf extract of Azadirachta indica; ALAO, aqueous leaf extract of Corchorus olitorius; ALCA, aqueous leaf extract of Cassia alata; ALPA, aqueous leaf extract of Phyllanthus amarus; ALVA, aqueous leaf extract of Vernonia amygdalina; ABAO, aqueous bark extract of Anacardium occidentale
Lipid peroxidation inhibitory activity of the aqueous extracts of the plants
The result in Fig. 2 revealed that the aqueous extracts of the selected plants demonstrated increased lipid peroxidation inhibitory activity with increase in the concentration of the extracts. However, at 800 mg/ml, ABAO (aqueous bark extract of Anacardium occidentale) exhibited highest lipid peroxidation inhibitory activity when compared with other extracts. Similarly, ABAO displayed higher inhibitory activity than ascorbic acid when compared with the standard antioxidants used. In all, gallic acid has the highest activity. This study revealed that the extracts could mitigate the lipid peroxidation chain reactions and prevent oxidation of cellular macromolecules.
Fig. 2.
Lipid peroxidation inhibitory activity of aqueous extracts of selected medicinal plants. ALAI, aqueous leaf extract of Azadirachta indica; ALAO, aqueous leaf extract of Corchorus olitorius; ALCA, aqueous leaf extract of Cassia alata; ALPA, aqueous leaf extract of Phyllanthus amarus; ALVA, aqueous leaf extract of Vernonia amygdalina; ABAO, aqueous bark extract of Anacardium occidentale
Ferric reducing power of the aqueous extracts of the plants
The result in Fig. 3 revealed the ferric reducing power of different concentrations of the aqueous extracts of the selected plants compared with standard antioxidants (ascorbic and tannic acids). At 800 mg/ml, all the extracts expect ALAO (aqueous leaf extract of Corchorus olitorius) displayed higher ferric radical reducing power than ascorbic acid when compared with standard antioxidants used. Of note, ABAO exhibited the highest ferric reducing power activity at 800 mg/mL among all the plant extracts used in the study.
Fig. 3.
Free radical reducing power of aqueous extracts of selected medicinal plants. ALAI, aqueous leaf extract of Azadirachta indica; ALAO, aqueous leaf extract of Corchorus olitorius; ALCA, aqueous leaf extract of Cassia alata; ALPA, aqueous leaf extract of Phyllanthus amarus; ALVA, aqueous leaf extract of Vernonia amygdalina; ABAO, aqueous bark extract of Anacardium occidentale
Hydroxyl radical scavenging ability of the aqueous extracts of the plants
Results in Fig. 4 showed that at 800 mg/ml, ALAI (aqueous leaf extract of Azadirachta indica), ALCA (aqueous leaf extract of Cassia alata), ALVA (aqueous leaf extract of Vernonia amygdalina), and ABAO (aqueous bark extract of Anacardium occidentale) have higher potent hydroxyl radical scavenging ability than ascorbic acid when compared with the standard antioxidants used. At all the tested concentrations, all the aqueous plant extracts displayed high hydroxyl radical scavenging ability with increase in their concentrations suggesting the extracts as potent source of antioxidant phytochemicals.
Fig. 4.
Hydroxyl radical scavenging activity of aqueous extracts of selected medicinal plants. ALAI, aqueous leaf extract of Azadirachta indica; ALAO, aqueous leaf extract of Corchorus olitorius; ALCA, aqueous leaf extract of Cassia alata; ALPA, aqueous leaf extract of Phyllanthus amarus; ALVA, aqueous leaf extract of Vernonia amygdalina; ABAO, aqueous bark extract of Anacardium occidentale
DPPH inhibitory activity of the aqueous extracts of the plants
The result in Fig. 5 shows that all the aqueous extracts of the selected medicinal plants used in this study elicited impressive DPPH radical scavenging activities when compared with standard antioxidants: gallic acid and ascorbic acid. At 800 mg/ml, ALVA (aqueous leaf extract of Vernonia amygdalina) exhibited the highest DPPH scavenging activity among all the extract used, followed by ABAO (aqueous bark extract of Anacardium occidentale) and then ALCA (aqueous leaf extract of Cassia alata) and ALPA. Although none of the extracts exhibited higher DPPH radical scavenging activity than the antioxidant standards used, their activity is significant, and this result depicted that they have substantial DPPH radical scavenging activity as the standard antioxidants.
Fig. 5.
DPPH radical scavenging activity of aqueous extracts of selected medicinal plants. ALAI, aqueous leaf extract of Azadirachta indica; ALAO, aqueous leaf extract of Corchorus olitorius; ALCA, aqueous leaf extract of Cassia alata; ALPA, aqueous leaf extract of Phyllanthus amarus; ALVA, aqueous leaf extract of Vernonia amygdalina; ABAO, aqueous bark extract of Anacardium occidentale
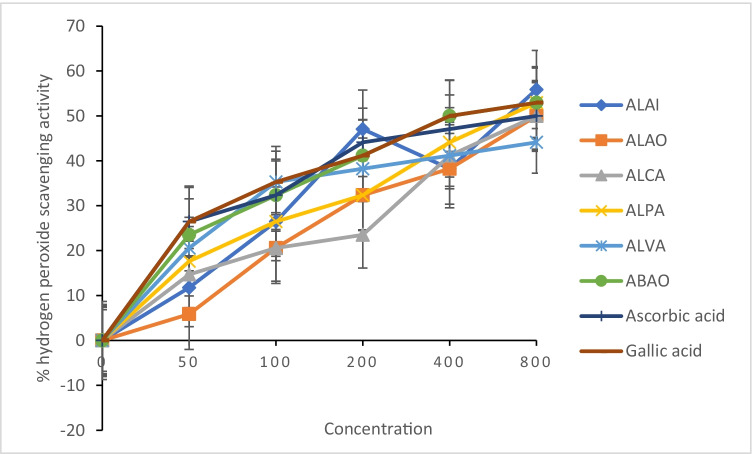
Hydrogen peroxide inhibitory activity of the aqueous extracts of the plants
Hydrogen peroxide scavenging activity of all the aqueous extracts of the plants used in this study was presented in Fig. 6 as compared with the standard antioxidants (ascorbic and gallic acids). At tested concentrations, all the aqueous extracts efficiently scavenged hydrogen peroxide. Interestingly, at 800 mg/ml, ALAI exhibited the highest hydrogen peroxide scavenging activity when compared with the standard antioxidants used and other extracts. Similarly, all the extracts expect ALVA displayed higher hydrogen peroxide scavenging activity than ascorbic acid. This result indicated that all the extracts could play beneficial role in mitigating pathological conditions mediated by oxidative stress.
Fig. 6.
Hydrogen peroxide scavenging activity of aqueous extracts of selected medicinal plants. ALAI, aqueous leaf extract of Azadirachta indica; ALAO, aqueous leaf extract of Corchorus olitorius; ALCA, aqueous leaf extract of Cassia alata; ALPA, aqueous leaf extract of Phyllanthus amarus; ALVA, aqueous leaf extract of Vernonia amygdalina; ABAO, aqueous bark extract of Anacardium occidentale
Phytochemical constituents and GCMS analysis of the extract
The preliminary qualitative phytochemical screening of extracts of the selected tropical plants used in this study revealed that the extracts are rich in phenol, flavonoids, coumarins, saponin, and alkaloids. The secondary metabolites profile of each of the extracts was detailed in Tables 1, 2, 3, 4, 5 and 6 as analysed using NIST 11 (Figs. 7, 8, 9, 10, 11 and 12). Some of the identified phytochemicals in the extracts have been reported to have antiapoptotic, antioxidant, anti-inflammatory, and cytoprotective effects which are essential to mitigate inflammation, oxidative stress, and toxicity effects mediated by Coronavirus-2.
Table 1.
GC–MS analysis of aqueous leaf of Azadirachta indica
| S/N | Compound | Canonical SMILES | PubChem CID |
|---|---|---|---|
| 1 | Ethyl acetate | CCOC(= O)C | 8857 |
| 2 | 2-Propenal | C = CC = O | 7847 |
| 2-Propen-1-amine | C = CCN | 7853 | |
| 3 | Acetic acid, butyl ester | CCCCOC(= O)C | 31,272 |
| 4 | 3-Hexanone, 2-methyl- | CCCC(= O)C(C)C | 23,847 |
| 3,4-Dimethylpent-2-en-1-ol | CC(C)C(= CCO)C | 5,366,252 | |
| 1,1-Di(isobutyl)acetone | CC(C)CC(CC(C)C)C(= O)C | 551,338 | |
| 5 | Ethylbenzene | CCC1 = CC = CC = C1 | 7500 |
| o-Xylene | CC1 = CC = CC = C1C | 7237 | |
| 6 | n-Butyl ether | CCCCOCCCC | 8909 |
| Propanoic acid, 2-methylpropyl ester | CCC(= O)OCC(C)C | 10,895 | |
| n-Butyl ether | CCCCOCCCC | 8909 | |
| 7 | Ethane, 1-chloro-2-isocyanato- | C(CCl)N = C = O | 16,035 |
| 1-[N-Aziridyl]heptene-3 | CCCC = CCCN1CC1 | 5,364,877 | |
| 1-Heptene, 2-methyl- | CCCCCC(= C)C | 27,519 | |
| 8 | 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | CC1(CC(C2 = C1C = CC(= C2)OC)(C)C3 = CC = C(C = C3)OC)C | 624,568 |
|
1-[2,4-Bis(trimethylsiloxy)phenyl] -2-[(4-trimethylsiloxy)phenyl]prop an-1-one |
|||
| Chloromethyl octyl ether | CCCCCCCCOCCl | 534,743 | |
| 9 | Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 |
| Chloroacetic acid, 2-methylpentylester | |||
| Cyclohexanol, 1R-4-acetamido-2,3-cis-epoxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 | |
| 10 | Butane, 2,2′-thiobis- | C(CCO)CO.C(CSCCO)O | 44,153,764 |
| 1,6-Anhydro-2,3-dideoxy-.beta.-D-erythro-hexopyranose | C1CC2OCC(C1O)O2 | 560,102 | |
| Boronic acid, ethyl-, bis(2-mercaptoethyl ester) | C1CC2OCC(C1O)O2 | 560,102 | |
| 11 | Silanol, trimethyl-, propanoate | CCC(= O)O[Si](C)(C)C | 519,321 |
| .beta.-D-Ribopyranoside, methyl 2, 3,4-tri-O-methyl- | COC1COC(C(C1OC)OC)OC | 21,140,439 | |
| Hexadecanoic acid, 3,7,11,15-tetramethyl-, methyl ester | CC(C)CCCC(C)CCCC(C)CCCC(C)CC(= O)OC | 568,163 | |
| 12 | N1,N1-Dimethyl-N2-(1-phenyl–ethyl) -ethane-1,2-diamine | CC(C1 = CC = CC = C1)NCCN(C)C | 547,403 |
| 2-Nonanone | CCCCCCCC(= O)C | 13,187 | |
| 13 | 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl ester | CCOC(= O)C1 = C(C(= C(N1)C)C2C(= C(OC3 = C2C(= O)CCC3)N)C#N)C | 605,143 |
| Heptasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13-tetradecamethyl- | C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,088 | |
|
1H-Indole-2-carboxylic acid, 6-(4-ethoxyphenyl)-3-methyl-4-oxo-4,5,6 ,7-tetrahydro-, isopropyl ester |
CCOC1 = CC = C(C = C1)C2CC3 = C(C(= C(N3)C(= O)OC(C)C)C)C(= O)C2 | 4,916,205 | |
| 14 | Octasiloxane, 1,1,3,3,5,5,7,7,9,9, 11,11,13,13,15,15-hexadecamethyl- | C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,087 |
| Heptasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13-tetradecamethyl- | C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,088 | |
|
Furo[2′,3′:4,5]thiazolo[3,2-g]purine-8-methanol, 4-amino-6.alpha.,7, 8,9a-tetrahydro-7-hydroxy-, [6aS-( 6a.alpha.,7.alpha.,8.beta.,9a.alph a.)]- |
|||
| 15 | Dodecanoic acid | CCCCCCCCCCCC(= O)O | 3893 |
| Methyl 6-O-[1-methylpropyl]-.beta. -d-galactopyranoside | CCC(C)OCC1C(C(C(C(O1)OC)O)O)O | 22,213,632 | |
| Nonanoic acid | CCCCCCCCC(= O)O | 8158 | |
| 16 | n-Decanoic acid | CCCCCCCCCC(= O)O | 2969 |
| Trisilane | C[Si](C)(C)[Si](C)(C)[Si](C)(C)C1 = CC = CC2 = CC = CC = C21 | 142,270 | |
| 4a-Methyl-6,8-dioxa-3-thia-bicyclo(3,2,1)octane | 50,469,286 | ||
| 17 | Butane, 1,1-dibutoxy- | CCCCOC(CCC)OCCCC | 22,210 |
| 1,1-Diisobutoxy-isobutane | CC(C)COC(C(C)C)OCC(C)C | 545,267 | |
| 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 | |
| 18 | Undec-10-ynoic acid | C#CCCCCCCCCC(= O)O | 31,039 |
| 19 | 1-Pentadecyne | CCCCCCCCCCCCCC#C | 69,825 |
| 20 | Spirohexan-4-one, 5,5-dimethyl- | CC1(CC2(C1 = O)CC2)C | 534,214 |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 | |
| 21 | Spirohexan-4-one, 5,5-dimethyl- | CC1(CC2(C1 = O)CC2)C | 534,214 |
| 1-Methoxymethoxy-oct-2-yne | CCCCCC#CCOCOC | 542,329 | |
| Thioctic acid | C1CSSC1CCCCC(= O)O | 864 | |
| 22 | 9,12-Octadecadienoic acid (Z,Z)- | CCCCCC = CCC = CCCCCCCCC(= O)O | 5,280,450 |
| 23 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 24 | Chloroacetic acid, 1-cyclopentylet hyl ester | CC(C1CCCC1)OC(= O)CCl | 543,343 |
| 25 | (4-Methoxymethoxy-hex-5-ynylidene) -cyclohexane | COCOC(CCC = C1CCCCC1)C#C | 542,331 |
| 26 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 3-Hydroxy-3-trifluoromethyl-2-oxa-spiro[5.5]undecan-5-one | C1CCC2(CC1)COC(CC2 = O)(C(F)(F)F)O | 557,008 | |
| 27 | Cyclohexanol, 2-(2-propynyloxy)-, trans- | C#CCOC1CCCCC1O | 12,522,342 |
| 5-Methoxymethoxyhexa-2,3-diene | CC = C = CC(C)OCOC | 542,425 | |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| 28 | Spirohexan-4-one, 5,5-dimethyl- | CC1(CC2(C1 = O)CC2)C | 534,214 |
| 3-Hydroxy-3-trifluoromethyl-2-oxa- spiro[5.5]undecan-5-one | C1CCC2(CC1)COC(CC2 = O)(C(F)(F)F)O | 557,008 | |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| 29 | 9,12-Octadecadienoic acid (Z,Z)- | CCCCCC = CCC = CCCCCCCCC(= O)O | 5,280,450 |
| 30 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| 1-Methoxymethoxy-oct-2-yne | CCCCCC#CCOCOC | 542,329 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 31 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| Spirohexan-4-one, 5,5-dimethyl- | CC1(CC2(C1 = O)CC2)C | 534,214 | |
| Cyclopentaneethanol, 2 (hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 32 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| 3-Hydroxy-3-trifluoromethyl-2-oxa- spiro[5.5]undecan-5-one | C1CCC2(CC1)COC(CC2 = O)(C(F)(F)F)O | 557,008 | |
| Chloroacetic acid, 1-cyclopentylethyl ester | CC(C1CCCC1)OC(= O)CCl | 543,343 | |
| 33 | Spirohexan-4-one, 5,5-dimethyl- | CC1(CC2(C1 = O)CC2)C | 534,214 |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 34 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
Table 2.
GC–MS analysis of aqueous leaf of Corchorus olitorius
| S/N | Compound | Canonical SMILES | PubChem CID |
|---|---|---|---|
| 1 | Ethyl acetate | CCOC(= O)C | 8857 |
| Acetic acid, pentyl ester | CCCCCOC(= O)C | 12,348 | |
| 2 | 2-Propenal | C = CC = O | 7847 |
| 2-Propen-1-amine | C = CCN | 7853 | |
| 3 | 2-Propenal | C = CC = O | 7847 |
| 2-Propen-1-amine | C = CCN | 7853 | |
| 4 | Acetic acid, butyl ester | CCCCOC(= O)C | 31,272 |
| 5 | 3-Hexanone, 2-methyl- | CCCC(= O)C(C)C | 23,847 |
| Butyric acid, 2,2-dimethyl-, vinyl ester | CCC(C)(C)C(= O)OC = C | 537,729 | |
| 6 | o-Xylene | CC1 = CC = CC = C1C | 7237 |
| Benzene, 1,3-dimethyl- | CC1 = CC(= CC = C1)C | 7929 | |
| 7 | n-Butyl ether | CCCCOCCCC | 8909 |
| Formic acid, 3,3-dimethylbut-2-yl ester | CC(C(C)(C)C)OC = O | 54,488,145 | |
| 8 | 4-Ethyl-4-methyl-5-methylene-[1,3] dioxolan-2-one | CCC1(C(= C)OC(= O)O1)C | 544,710 |
| 1,1-Cyclohexanedicarbonitrile | C1CCC(CC1)(C#N)C#N | 544,561 | |
| Ethane, 1-chloro-2-isocyanato- | C(CCl)N = C = O | 16,035 | |
| 9 | Acetamide, N-cyclohexyl- | CC(= O)NC1CCCCC1 | 14,301 |
| Acetamide, N-(4-hydroxycyclohexyl) -, cis- | CC(= O)NC1CCC(CC1)O | 90,074 | |
| L-Lyxose | C(C(C(C(C = O)O)O)O)O | 644,176 | |
| 10 | 12,12-Dimethoxydodecanoic acid, methyl ester | COC(CCCCCCCCCCC(= O)OC)OC | 554,421 |
| Hexadecane, 1,1-dimethoxy- | CCCCCCCCCCCCCCCC(OC)OC | 76,037 | |
| 1-Propene, 3,3-dichloro- | C = CC(Cl)Cl | 11,244 | |
| 11 | Isopropyl isothiocyanate | CC(C)N = C = S | 75,263 |
| Tridecyl (E)-2-methylbut-2-enoate | CCCCCCCCCCCCCOC(= O)C(= CC)C | 91,701,893 | |
| 3-Phenylpropionic acid, 4-methoxy- 2-methylbutyl ester | CC(CCOC)COC(= O)CCC1 = CC = CC = C1 | 91,702,047 | |
| 12 | Hexasiloxane, 11,11-dodecamethyl- | C[Si](C)(O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)Cl)O[Si](C)(C)O[Si](C)(C)Cl | 553,580 |
|
Octasiloxane, 1,1,3,3,5,5,7,7,9,9, 11,11,13,13,15,15 hexadecamethyl- |
C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,087 | |
| 9-Bromononanoic acid | C(CCCCBr)CCCC(= O)O | 548,221 | |
| 13 | Hexasiloxane, tetradecamethyl- | C[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)C | 7875 |
| Octasiloxane, 1,1,3,3,5,5,7,7,9,9, 11,11,13,13,15,15-hexadecamethyl- | C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,087 | |
| 1-Butyl-3,4-dihydroxy- | CCCCCNC(= O)C1C(C(CN1CCCC)O)O | 6,420,934 | |
| pyrrolidine- 2,5-dione | C1C(C(= O)N(C1 = O)C2 = CC = C(C = C2)F)SCCN | 2,756,427 | |
| 14 | 11-Bromoundecanoic acid | C(CCCCCBr)CCCCC(= O)O | 17,812 |
| 2-Ethylacridine | CCC1 = CC2 = CC3 = CC = CC = C3N = C2C = C1 | 610,161 | |
| N-Cyclododecylacetamide | CC(= O)NC1CCCCCCCCCCC1 | 548,224 | |
| 15 | Octadecanoic acid | CCCCCCCCCCCCCCCCCC(= O)O | 5281 |
| Polygalitol | C1C(C(C(C(O1)CO)O)O)O | 64,960 | |
| Acetamide, N-cyclohexyl-2-[(2-furanylmethyl)thio]- | C1CCC(CC1)NC(= O)CSCC2 = CC = CO2 | 5,298,618 | |
| 16 | Butane, 1,1-dibutoxy- | CCCCOC(CCC)OCCCC | 22,210 |
| 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 | |
| 1,1-Diisobutoxy-isobutane | CC(C)COC(C(C)C)OCC(C)C | 545,267 | |
| 17 | 1-Butyl-3,4-dihydroxy-pyrrolidine- 2,5-dione | CCCCN1C(= O)C(C(C1 = O)O)O | 6,420,934 |
| L-Galactose, 6-deoxy- | CC(C(C(C(C = O)O)O)O)O | 3,034,656 | |
| Nonanoic acid | CCCCCCCCC(= O)O | 8158 | |
| 18 | 9,12-Octadecadienoic acid (Z,Z)- | CCCCCC = CCC = CCCCCCCCC(= O)O | 5,280,450 |
| 9-Octadecyne | CCCCCCCCC#CCCCCCCCC | 141,998 | |
| Undec-10-ynoic acid | C#CCCCCCCCCC(= O)O | 31,039 | |
| 19 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 | |
| Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 | |
| 20 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| Thioctic acid | C1CSSC1CCCCC(= O)O | 864 | |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 | |
| 21 | Spirohexan-4-one, 5,5-dimethyl- | CC1(CC2(C1 = O)CC2)C | 534,214 |
| 1-Methoxymethoxy-oct-2-yne | CCCCCC#CCOCOC | 542,329 | |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| 22 | Thioctic acid | C1CSSC1CCCCC(= O)O | 864 |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| (4-Methoxymethoxy-hex-5-ynylidene -cyclohexane | COCOC(CCC = C1CCCCC1)C#C | 542,331 | |
| 23 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
|
1-Naphthalenemethanol, decahydro-5 -(5-hydroxy-3-methyl-3-pentenyl)-1 ,4a-dimethyl-6-methylene-, [1S-[1.alpha.,4a.alpha.,5.alpha.(E),8a.beta.]]- |
CC(= CCO)CCC1C(= C)CCC2C1(CCCC2(C)CO)C | 5,316,778 | |
| 3-Hydroxy-3-trifluoromethyl-2-oxa spiro[5.5]undecan-5-one | C1CCC2(CC1)COC(CC2 = O)(C(F)(F)F)O | 557,008 | |
| 24 |
1-Naphthalenemethanol, decahydro-5 -(5-hydroxy-3-methyl-3-pentenyl)-1 ,4a-dimethyl-6-methylene-, [1S-[1.alpha.,4a.alpha.,5.alpha.(E),8a.be ta.]]- |
CC(= CCO)CCC1C(= C)CCC2C1(CCCC2(C)CO)C | 5,316,778 |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 | |
| 25 | Cyclobutanone, 2-methyl-2-oxiranyl | CC1(CCC1 = O)C2CO2 | 534,052 |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 26 | Borolane, 3-ethyl-4-methyl-1,2,2-tris(1-methylethyl)- | B1(CC(C(C1(C(C)C)C(C)C)CC)C)C(C)C | 535,085 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 27 | 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
| Cyclohexanol, 2-(2-propynyloxy)-, trans- | C#CCOC1CCCCC1O | 12,522,342 | |
| Spiro[2.3]hexan-4-one, 5,5-diethyl | CCC1(CC2(C1 = O)CC2)CC | 534,343 | |
| 28 |
3- Oxatricyclo[4.2.0.0(2,4)]octan -one |
C1C2C(CC2 = O)C3C1O3 | 556,833 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| Cyclododecanol, 1-aminomethyl- | C1CCCCCC(CCCCC1)(CN)O | 533,851 | |
| 29 | Cyclobutanone, 2-methyl-2-oxiranyl | CC1(CCC1 = O)C2CO2 | 534,052 |
| Spiro[2.3]hexan-4-one, 5,5-diethyl | CCC1(CC2(C1 = O)CC2)CC | 534,343 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 30 | Cyclohexanol, 2-(2-propynyloxy)-, trans- | C#CCOC1CCCCC1O | 12,522,342 |
| 8-Azabicyclo[3.2.1]octane-2-carboxylic acid, 3-hydroxy-8-methyl-, (2-endo,3-exo)- | CN1C2CC1C(C(C(C2)O)C(= O)[O-])O | 54,613,783 | |
| Cyclobutanone, 2-methyl-2-oxiranyl | CC1(CCC1 = O)C2CO2 | 534,052 | |
| 31 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| Spirohexan-4-one, 5,5-dimethyl- | CCC1(CC2(C1 = O)CC2)CC | 534,343 | |
| 5-Methoxymethoxyhexa-2,3-diene | CC = C = CC(C)OCOC | 542,425 | |
| 32 | Spirohexan-4-one, 5,5-dimethyl- | CCC1(CC2(C1 = O)CC2)CC | 534,343 |
| Cyclopentaneethanol, 2-(hydroxyl methyl)-.beta.,3-dimethyl- | |||
| 5-Methoxymethoxyhexa-2,3-diene | CC = C = CC(C)OCOC | 542,425 | |
| 33 | Borolane, 3-ethyl-4-methyl-1,2,2-tris(1-methylethyl)- | B1(CC(C(C1(C(C)C)C(C)C)CC)C)C(C)C | 535,085 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 | |
| Thioctic acid | C1CSSC1CCCCC(= O)O | 864 | |
| 34 | 5-Methoxymethoxyhexa-2,3-diene | CC = C = CC(C)OCOC | 542,425 |
| Cyclohexanol, 2-(2-propynyloxy)-, trans- | C#CCOC1CCCCC1O | 12,522,342 | |
| Ethane, 1,2-dichloro-1,1,2-trifluoro- | C(C(F)(F)Cl)(F)Cl | 9631 | |
| 35 | 5-Methoxymethoxyhexa-2,3-diene | CC = C = CC(C)OCOC | 542,425 |
| Cyclohexanone, 2-(2 propenyl)- | CC(= CC1CCCCC1 = O)C | 592,361 | |
| 3-Hydroxy-3-trifluoromethyl-2-oxa-spiro[5.5]undecan-5-one | C1CCC2(CC1)COC(CC2 = O)(C(F)(F)F)O | 557,008 | |
| 36 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| Spirohexan-4-one, 5,5-dimethyl- | CCC1(CC2(C1 = O)CC2)CC | 534,343 | |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC23C(C1)CCC(= O)C2O3 | 548,904 |
Table 3.
GC–MS analysis of aqueous leaf of Cassia alata
| S/N | Compound | Canonical SMILES | PubChem CID |
|---|---|---|---|
| 1 | Acetic acid, butyl ester | CCCCOC(= O)C | 31,272 |
| 2 | 3-Hexanone, 2-methyl- | CCCC(= O)C(C)C | 23,847 |
| Acetaldehyde, butylhydrazone | CCCCNN = CC | 9,601,789 | |
| Butyric acid, 2,2-dimethyl-, vinyl ester | CCC(C)(C)C(= O)OC = C | 537,729 | |
| 3 | o-Xylene | CC1 = CC = CC = C1C | 7237 |
| 1-Oxa-4-azaspiro[4.5]decan-4-oxyl, 3,3-dimethyl-8-oxo- | CC1(COC2([N +]1 = O)CCC(= O)CC2)C | 6,420,654 | |
| 1,3,2-Dioxaborinane, 2-ethyl-5,5-dimethyl- | B1(OCC(CO1)(C)C)CC | 544,611 | |
| 4 | n-Butyl ether | CCCCOCCCC | 8909 |
| Propanoic acid, 2-methylpropyl ester | CCC(= O)OCC(C)C | 10,895 | |
| 5 | Pyrazol-3(2H)-one, 4-(2-furfurylidenamino)-1,5-dimethyl-2-phenyl- | CC1 = C(C(= O)N(N1C)C2 = CC = CC = C2)N = CC3 = CC = CO3 | 624,533 |
| Cyclohexaneacetic acid, butyl este | CCCCOC(= O)CC1CCCCC1 | 251,210 | |
|
Furo[2′,3′:4,5]thiazolo[3,2-g]purine-8-methanol, 4-amino-6.alpha.,7,8,9a-tetrahydro-7-hydroxy-, [6aS-(6a.alpha.,7.alpha.,8.beta.,9a.alph a.)]- |
|||
| 6 | 6-Azaestra-1,3,5(10),6,8-pentaen-1,7-one, 3-methoxy- | ||
| Pyrazol-3(2H)-one, 4-(2-furfurylidenamino)-1,5-dimethyl-2-phenyl- | CC1 = C(C(= O)N(N1C)C2 = CC = CC = C2)N = CC3 = CC = CO3 | 624,533 | |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | CC1(CC(C2 = C1C = CC(= C2)OC)(C)C3 = CC = C(C = C3)OC)C | 624,568 | |
| 7 | Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 |
| Cyclohexaneacetic acid, butyl este | CCCCOC(= O)CC1CCCCC1 | 251,210 | |
| Cyclopentaneundecanoic acid | C1CCC(C1)CCCCCCCCCCC(= O)O | 534,549 | |
| 8 | Hexyl 2-hydroxyethyl sulfide | CCCCCCSCCO | 90,519 |
| D-Arabinitol | C(C(C(C(CO)O)O)O)O | 94,154 | |
| Ribitol | C(C(C(C(CO)O)O)O)O | 6912 | |
| 9 | Silanol, trimethyl-, propanoate | CCC(= O)O[Si](C)(C)C | 519,321 |
|
6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsu lfanyl-ethyl ester |
|||
| 2-Octanol, 8,8-dimethoxy-2,6-dimethyl- | CC(CCCC(C)(C)O)CC(OC)OC | 31,272 | |
| 10 | Ethanol, 2-phenoxy-, propanoate | CCC(= O)OCCOC1 = CC = CC = C1 | 31,954 |
| Tridecyl (E)-2-methylbut-2-enoate | CCCCCCCCCCCCCOC(= O)C(= CC)C | 91,701,893 | |
| 11 | Butane, 1,1-dibutoxy- | CCCCOC(CCC)OCCCC | 22,210 |
| 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 |
Table 4.
GC–MS analysis of aqueous leaf of Phyllanthus amarus
| S/N | Compound | Canonical SMILES | PubChem CID |
|---|---|---|---|
| 1 | Acetic acid, butyl ester | CCCCOC(= O)C | 31,272 |
| 2 | 3-Hexanone, 2-methyl- | CCCC(= O)C(C)C | 23,847 |
| Butyric acid, 2,2-dimethyl-, vinylester | CCC(C)(C)C(= O)OC = C | 537,729 | |
| 2-Methylvaleroyl chloride | CCCC(C)C(= O)Cl | 107,385 | |
| 3 | o-Xylene | CC1 = CC = CC = C1C | 7237 |
| 1-Hexene, 3,4-dimethyl- | CCC(C)C(C)C = C | 140,134 | |
| 2H-Pyran-2-one, tetrahydro-5,6-dimethyl-, trans- | CC1CCC(= O)OC1C | 11,029,861 | |
| 4 | n-Butyl ether | CCCCOCCCC | 8909 |
| Formic acid, 3,3-dimethylbut-2-ylester | CC(C(C)(C)C)OC = O | 54,488,145 | |
| 5 | Pentane, 1-bromo-3,4-dimethyl- | CC(C)C(C)CCBr | 544,665 |
| Butanoic acid, butyl ester | CCCCOC(= O)CCC | 7983 | |
| Cyclododecylamine | C1CCCCCC(CCCCC1)N | 2897 | |
| 6 | Pentane, 1-bromo-3,4-dimethyl- | CC(C)C(C)CCBr | 544,665 |
| Pentane, 2,3-dimethyl- | CCC(C)C(C)C | 11,260 | |
| 2,4-Dipropyl-5-ethyl-1,3-dioxane | CCCC1C(COC(O1)CCC)CC | 221,917 | |
| 7 | n-Butyl isobutyl sulfide | CCCCSCC(C)C | 525,418 |
| 1,6-Anhydro-2,3-dideoxy-.beta.-D-erythro-hexopyranose | C1CC2OCC(C1O)O2 | 560,102 | |
| Decanoic acid, propyl ester | CCCCCCCCCC(= O)OCCC | 121,739 | |
| 8 | 6-Bromohexanoic acid, hexyl ester | CCCCCCOC(= O)CCCCCBr | 543,783 |
| 1,3-Hexanediol, 2-ethyl- | CCCC(C(CC)CO)O | 7211 | |
| 1-Undecene, 2-methyl- | CCCCCCCCCC(= C)C | 87,689 | |
| 9 | 1,3-Dioxane, 2-ethyl-5-methyl- | CCC1OCC(CO1)C | 141,976 |
| Silanol, trimethyl-, propanoate | CCC(= O)O[Si](C)(C)C | 519,321 | |
| .beta.-D-Ribopyranoside, methyl 2, 3,4-tri-O-methyl- | COC1COC(C(C1OC)OC)OC | 21,140,439 | |
| 10 | Propane, 1-isothiocyanato- | CCCN = C = S | 69,403 |
| Butane, 1,1′-[ethylidenebis(oxy)]bis- | CCC(C)COC(C)OCC(C)CC | 551,340 | |
| 11 | 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 |
| Para methadione | CCC1(C(= O)N(C(= O)O1)C)C | 8280 | |
| 12 | Butane, 1,1-dibutoxy- | CCCCOC(CCC)OCCCC | 22,210 |
| 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 | |
| Neopentyl isothiocyanate | CC(C)(C)CN = C = S | 136,393 |
Table 5.
GC–MS analysis of aqueous leaf of Vernonia amygdalina
| S/N | Compound | Canonical SMILES | PubChem CID |
|---|---|---|---|
| 1 | Ethyl acetate | CCOC(= O)C | 8857 |
| 2 | 2-Propenal | C = CC = O | 7847 |
| 2-Propen-1-amine | C = CCN | 7853 | |
| 3 | 2-Propenal | C = CC = O | 7847 |
| Aziridine, 2-methyl- | CC1CN1 | 6377 | |
| 4 | Acetic acid, butyl ester | CCCCOC(= O)C | 31,272 |
| 5 | Ethanamine, N-pentylidene- | CCCCC = NCC | 544,805 |
| 2-Propanone, propylhydrazone | CCCNN = C(C)C | 139,018 | |
| Oxirane, (1-methylethyl)- | CC(C)C1CO1 | 102,618 | |
| 6 | Pyrazol-3(2H)-one, 4-(5-hydroxymethylfurfur-2-ylidenamino)-1,5-dimethyl-2-phenyl- | CC1 = C(C(= O)N(N1C)C2 = CC = CC = C2)N = CC3 = CC = C(O3)CO | 544,706 |
| Ethylbenzene | CCC1 = CC = CC = C1 | 7500 | |
| 7 | n-Butyl ether | CCCCOCCCC | 8909 |
|
Formic acid, 3,3-dimethylbut-2-yl ester |
CC(C(C)(C)C)OC = O | 54,488,145 | |
| 8 | Ethane, 1-chloro-2-isocyanato- | C(CCl)N = C = O | 16,035 |
| 1-Hexene, 3,4-dimethyl- | CCC(C)C(C)C = C | 140,134 | |
| 3,4,5-Trimethyldihydrofuran-2-one | CC1C(C(= O)OC1C)C | 544,600 | |
| 9 | Cyclohexane acetic acid, butyleste | ||
| Acetamide, N-cyclohexyl- | CC(= O)NC1CCCCC1 | 14,301 | |
| Pentane, 1-bromo-3,4-dimethyl- | CC(C)C(C)CCBr | 544,665 | |
| 10 | N-Cyclododecylacetamide | CC(= O)NC1CCCCCCCCCCC1 | 548,224 |
| Cyclohexaneacetic acid, butyl este | |||
| Trisilane | C[Si](C)(C)[Si](C)(C)[Si](C)(C)C1 = CC = CC2 = CC = CC = C21 | 142,270 | |
| 11 | 1,6-Anhydro-2,3-dideoxy-.beta.-D-erythro-hexopyranose | C1CC2OCC(C1O)O2 | 560,102 |
| 2-Butenoic acid, butyl ester | CCCCOC(= O)C = CC | 5,366,039 | |
| Morpholine | C1COCCN1 | 8083 | |
| 12 | Dimethyl{bis[(3-methylbut-2-en-1-yl)oxy]}silane | CC(= CCO[Si](C)(C)OCC = C(C)C)C | 91,697,225 |
| 1-Propanol, 2,2-dimethyl-, acetate | CC(= O)OCC(C)(C)C | 13,552 | |
| Allyl(2-butoxy)dimethylsilane | CCC(C)O[Si](C)(C)CC = C | 554,670 | |
| 13 | Succinic acid, hexyl 3-oxobut-2-yl-ester | CCCCCCOC(= O)CCC(= O)OC(C)C(= O)C | 91,702,832 |
| .beta.-D-Xylopyranoside, methyl 2,3,4-tri-O-methyl- | COC1COC(C(C1OC)OC)OC2C(C(COC2OC)OC)OC | 91,726,683 | |
| Tridecyl (E)-2-methylbut-2-enoate | CCCCCCCCCCCCCOC(= O)C(= CC)C | 91,701,893 | |
| 14 | Thiazole, 2-ethyl-4,5-dihydro- | CCC1 = NCCS1 | 86,896 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 | |
| Cyclopentane, 1-methyl-2-(4-methylpentyl)-, trans- | CC1CCCC1CCCC(C)C | 6,432,631 | |
| 15 | Cyclopentaneundecanoic acid | C1CCC(C1)CCCCCCCCCCC(= O)O | 534,549 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 | |
| Pentadecanoic acid | CCCCCCCCCCCCCCC(= O)O | 13,849 | |
| 16 | Butane, 1,1-dibutoxy- | CCCCOC(CCC)OCCCC | 22,210 |
| 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 | |
| 17 | Dodecanoic acid | CCCCCCCCCCCC(= O)O | 3893 |
|
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
CC(C)(C)OC(= O)NC(CC1 = CC = CC = C1)CO | 2,733,675 | |
| 8-Chlorocapric acid | C(CCCC(= O)O)CCCCl | 548,250 | |
| 18 | Acetamide, N-cyclohexyl-2-[(2-furanylmethyl)thio]- | C1CCC(CC1)NC(= O)CSCC2 = CC = CO2 | 5,298,618 |
| n-Decanoic acid | CCCCCCCCCC(= O)O | 2969 | |
| Pentadecanoic acid | CCCCCCCCCCCCCCC(= O)O | 13,849 | |
| 19 | Acetamide, N-cyclohexyl-2-[(2-furanylmethyl)thio]- | C1CCC(CC1)NC(= O)CSCC2 = CC = CO2 | 5,298,618 |
| Propane, 1,2-dichloro-2-fluoro- | CCCCCCCCCCCCCCC(= O)O | 13,849 | |
| 1-Butyl-3,4-dihydroxy-pyrrolidine- 2,5-dione | CCCCN1C(= O)C(C(C1 = O)O)O | 6,420,934 | |
| 20 | 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| Ethanol, 2-bromo- | C(CBr)O | 10,898 | |
| 21 | Borolane, 3-ethyl-4-methyl-1,2,2-tris(1-methylethyl)- | B1(CC(C(C1(C(C)C)C(C)C)CC)C)C(C)C | 535,085 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 22 | Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 23 | Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 |
| 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 | |
| Cyclohexanone, 2-(1-mercapto-1-methylethyl)-5-methyl-, trans- | CC1CCC(C(= O)C1)C(C)(C)S | 6,951,713 | |
| 24 | Cyclohexanone, 2-(1-mercapto-1-methylethyl)-5-methyl-, trans- | CC1CCC(C(= O)C1)C(C)(C)S | 6,951,713 |
| Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 25 | 1,4-Cyclohexanedimethanamine | C1CC(CCC1CN)CN | 17,354 |
| Borolane, 3-ethyl-4-methyl-1,2,2-tris(1-methylethyl)- | B1(CC(C(C1(C(C)C)C(C)C)CC)C)C(C)C | 535,085 | |
| (4-Methoxymethoxy-hex-5-ynylidene)-cyclohexane | COCOC(CCC = C1CCCCC1)C#C | 542,331 | |
| 26 | Cyclobutanone, 2-methyl-2-oxiranyl | COCOC(CCC = C1CCCCC1)C#C | 542,331 |
| Borolane, 3-ethyl-4-methyl-1,2,2-tris(1-methylethyl)- | B1(CC(C(C1(C(C)C)C(C)C)CC)C)C(C)C | 535,085 | |
| Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 | |
| 27 | Cyclobutanone, 2-methyl-2-oxiranyl | COCOC(CCC = C1CCCCC1)C#C | 542,331 |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | C1C2C(CC2 = O)C3C1O3 | 556,833 | |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| 28 | Cyclobutanone, 2-methyl-2-oxiranyl | COCOC(CCC = C1CCCCC1)C#C | 542,331 |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | C1C2C(CC2 = O)C3C1O3 | 556,833 | |
| 29 | 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 |
| 2-Tetradecanol | CCCCCCCCCCCCC(C)O | 20,831 | |
| 2H-1,2,3-Triazol-4-amine, 2-cyclohexyl-5-nitro-, 1-oxide | 249,914,956 | ||
| 30 | 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | C1C2C(CC2 = O)C3C1O3 | 556,833 | |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 31 | Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 |
| Oxonin, 4,5,6,7-tetrahydro-, (Z,Z)32 | |||
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| 32 | Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC = COC = CC1 | 5,365,711 | |
| 33 | Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 |
| (4-Methoxymethoxy-hex-5-ynylidene)25-cyclohexane | COCOC(CCC = C1CCCCC1)C#C | 542,331 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
Table 6.
GC–MS analysis of aqueous bark of Anacardium occidentale
| S/N | Compound | Canonical SMILES | PubChem CID |
|---|---|---|---|
| 1 | Ethyl acetate | CCOC(= O)C | 8857 |
| 2 | 2-Propenal | C = CC = O | 7847 |
| 2-Propen-1-amine | C = CCN | 7853 | |
| 3 | Acetic acid, butyl ester | CCCCOC(= O)C | 31,272 |
| 4 | Ethanamine, N-pentylidene- | CCCCC = NCC | 544,805 |
| Acetaldehyde, butylhydrazone | CCCCNN = CC | 9,601,789 | |
| 2-Propanone, propylhydrazone | CCCNN = C(C)C | 139,018 | |
| 5 | p-Xylene | CC1 = CC = C(C = C1)C | 7809 |
| o-Xylene | CC1 = CC = CC = C1C | 7237 | |
| Ethylbenzene | CCC1 = CC = CC = C1 | 7500 | |
| 6 | n-Butyl ether | CCCCOCCCC | 8909 |
| Formic acid, 3,3-dimethylbut-2-yl ester | CC(C(C)(C)C)OC = O | 54,488,145 | |
| Ethanol, 2-butoxy- | CCCCOCCO | 8133 | |
| 7 | Pyrazol-3(2H)-one, 4-(5-hydroxymethylfurfur-2-ylidenamino)-1,5-dimethyl-2-phenyl- | CC1 = C(C(= O)N(N1C)C2 = CC = CC = C2)N = CC3 = CC = C(O3)CO | 544,706 |
| Ethane, 1-chloro-2-isocyanato- | C(CCl)N = C = O | 16,035 | |
| 1-Oxa-4-azaspiro[4.5]decan-4-oxyl, 3,3-dimethyl-8-oxo- | CC1(COC2([N +]1 = O)CCC(= O)CC2)C | 6,420,654 | |
| 8 | 1-Octene, 2-methyl- | CCCCCCC(= C)C | 78,335 |
| 1-Pentene, 5-chloro-4-(chloromethyl)-2,4-dimethyl- | CC(= C)CC(C)(CCl)CCl | 544,717 | |
| 4-t-Butylcyclohexylamine | CC(C)(C)C1CCC(CC1)N | 79,396 | |
| 9 | Butane, 2,2′-thiobis- | C(CCO)CO.C(CSCCO)O | 44,153,764 |
| Dimethylamine, N-(neopentyloxy)- | CC(C)(C)CON(C)C | 548,341 | |
| 3-Deoxy-d-mannitol | C(C(CO)O)C(C(CO)O)O | 560,035 | |
| 10 | Silane, trimethyl(2-methylpropoxy) | CC(C)CO[Si](C)(C)C | 519,538 |
| 1-Propanol, 2,2-dimethyl-, acetate | CC(= O)OCC(C)(C)C | 13,552 | |
| Propanoic acid, dimethyl(chloromethyl)silyl ester | CCC(= O)O[Si](C)(C)CCl | 554,674 | |
| 11 | 2-Butylthiazoline | CCCCC1 = NCCS1 | 206,597 |
| 4.beta.,5-Dimethyl-6,8-dioxa-3-thiabicyclo(3,2,1)octane 3-oxide | CC1C2(OCC(O2)CS1 = O)C | 538,769 | |
| Succinic acid, butyl decyl ester | CCCCCCCCCCOC(= O)CCC(= O)OCCCC | 91,701,657 | |
| 12 | 1-Pentadecene, 2-methyl- | CCCCCCCCCCCCCC(= C)C | 520,456 |
| Pyrido[2,3-d]pyrimidine, 4-phenyl- | C1 = CC = C(C = C1)C2 = C3C = CC = NC3 = NC = N2 | 610,177 | |
| 1-Benzenesulfonyl-1H-pyrrole | C1 = CC = C(C = C1)S(= O)(= O)N2C = CC = C2 | 140,146 | |
| 13 | Acetamide, N-(4-hydroxycyclohexyl) -, cis- | CC(= O)NC1CCC(CC1)O | 90,074 |
| Octasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyl- | C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,087 | |
| 1H-Indole, 5-methyl-2-phenyl- | CC1 = CC2 = C(C = C1)NC(= C2)C3 = CC = CC = C3 | 83,247 | |
| 14 | Thiazole, 2-ethyl-4,5-dihydro- | CCC1 = NCCS1 | 86,896 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 | |
| Cyclopentaneundecanoic acid | C1CCC(C1)CCCCCCCCCCC(= O)O | 534,549 | |
| 15 | Cyclopentaneundecanoic acid | C1CCC(C1)CCCCCCCCCCC(= O)O | 534,549 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | CC(= O)NC1CCC(C2C1O2)OC = O | 537,204 | |
| Decanoic acid, silver(1 +) salt | 319,368,662 | ||
| 16 | Butane, 1,1-dibutoxy- | CCCCOC(CCC)OCCCC | 22,210 |
| 1-Butoxy-1-isobutoxy-butane | CCCCOC(CCC)OCC(C)C | 545,190 | |
| Neopentyl isothiocyanate | CC(C)(C)CN = C = S | 136,393 | |
| 17 | Nonanoic acid | CCCCCCCCC(= O)O | 8158 |
| .alpha.-D-Glucopyranose, 4-O-.beta.-D-galactopyranosyl | C(C1C(C(C(C(O1)OC2C(OC(C(C2O)O)O)CO)O)O)O)O.O | 522,113 | |
|
n- Decanoic acid |
CCCCCCCCCC(= O)O | 2969 | |
| 18 | Tridecanoic acid | CCCCCCCCCCCCC(= O)O | 12,530 |
| Octasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyl- | C[Si](C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)(C)O[Si](C)C | 6,329,087 | |
| 19 | Z,E-7,11-Hexadecadien-1-yl acetate | CCCCC = CCCC = CCCCCCCOC(= O)C | 5,363,282 |
|
2(1H)-Naphthalenone, octahydro-4a- methyl-7-(1-methylethyl)-, (4a.alp ha.,7.beta.,8a.beta.) - |
|||
|
3,6-Epoxy-2H,8H-pyrimido[6,1-b][1,3]oxazocine-8,10(9H)-dione, 4-(ace tyloxy)-3,4,5,6-tetrahydro-11-meth yl-, [3R-(3.alpha.,4.beta.,6.alpha .)]- |
|||
| 20 | 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
| Chloroacetic acid, 1-cyclopentylethyl ester | CC(C1CCCC1)OC(= O)CCl | 543,343 | |
| Dodecanoic acid, 1,2,3-propanetriyl ester | CCCCCCCCCCCC(= O)OCC(COC(= O)CCCCCCCCCCC)OC(= O)CCCCCCCCCCC | 10,851 | |
| 21 | 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | C1CCC = COC = CC1 | 5,365,711 | |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 22 | Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| Spiro[2.3]hexan-4-one, 5,5-diethyl | CC1(CC2(C1 = O)CC2)C | 534,214 | |
| 23 | Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | C1C2C(CC2 = O)C3C1O3 | 556,833 | |
| Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 24 | Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 | |
| 25 | 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | C1C2C(CC2 = O)C3C1O3 | 556,833 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 26 | 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| Chloroacetic acid, 1-cyclopentylethyl ester | CC(C1CCCC1)OC(= O)CCl | 543,343 | |
| 27 | 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 |
| Cyclohexanone, 2-(2-propenyl)- | C = CCC1CCCCC1 = O | 78,944 | |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | C1C2C(CC2 = O)C3C1O3 | 556,833 | |
| 28 | Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| 2-Butynamide, N-methyl- | CC#CC(= O)NC | 549,138 | |
| 29 | Dodecanoic acid, 1,2,3-propanetriyl ester | CCCCCCCCCCCC(= O)OCC(COC(= O)CCCCCCCCCCC)OC(= O)CCCCCCCCCCC | 10,851 |
| [1,2,4]Triazolo[1,5-a]pyrimidin-5(4H)-one, 7-amino-4-methyl- | CN1C(= O)C = C(N2C1 = NC = N2)N | 16,766,882 | |
| 30 | Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 31 | Dodecanoic acid, 1,2,3-propanetriyl ester | CCCCCCCCCCCC(= O)OCC(COC(= O)CCCCCCCCCCC)OC(= O)CCCCCCCCCCC | 10,851 |
| Cyclododecanol, 1-aminomethyl- | C1CCCCCC(CCCCC1)(CN)O | 533,851 | |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 32 | Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | CC1CCC(C1CO)C(C)CO | 101,710 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | CCCCCCC(CCO)CCOC | 542,174 | |
| Cyclobutanone, 2-methyl-2-oxiranyl | CC1CCC(C1CO)C(C)CO | 101,710 | |
| 33 | Cyclododecanol, 1-aminomethyl- | C1CCCCCC(CCCCC1)(CN)O | 533,851 |
| 1H-Imidazole, 2-ethyl-4,5-dihydro- | CCC1 = NCCN1 | 13,590 | |
| 1-Phenyloxycarbonyl-7-pentyl-7-aza bicyclo[4.1.0]heptane |
Fig. 7.
GC–MS spectrum of aqueous leaf of Azadirachta indica
Fig. 8.
GC–MS spectrum of aqueous leaf of Corchorus olitorius
Fig. 9.
GC–MS spectrum of aqueous leaf of Cassia alata
Fig. 10.
GC–MS spectrum of aqueous leaf of Phyllanthus amarus
Fig. 11.
GC–MS spectrum of aqueous leaf of Vernonia amygdalina
Fig. 12.
GC–MS spectrum of aqueous bark of Anacardium occidentale
Molecular docking analysis
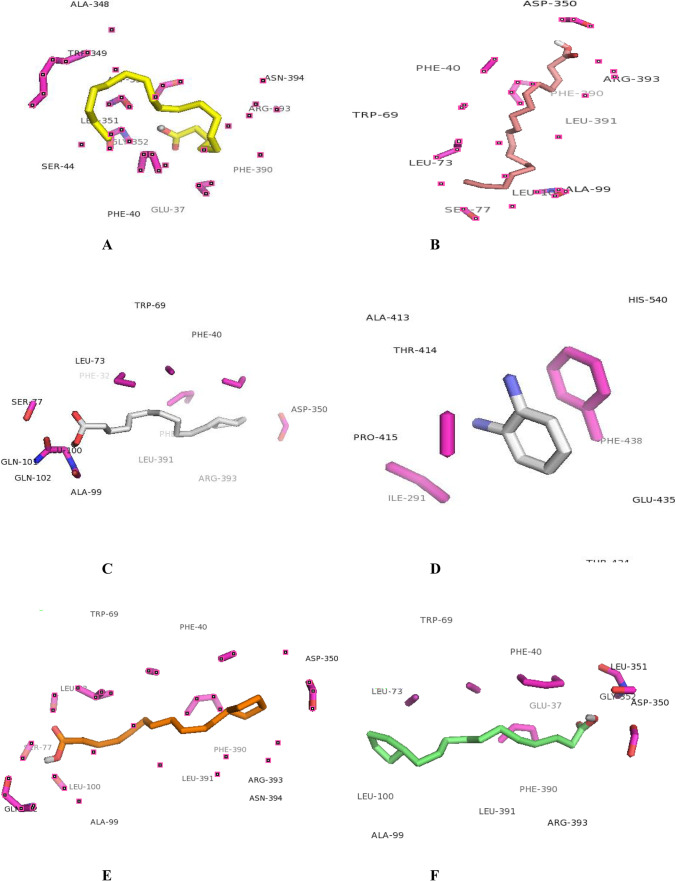
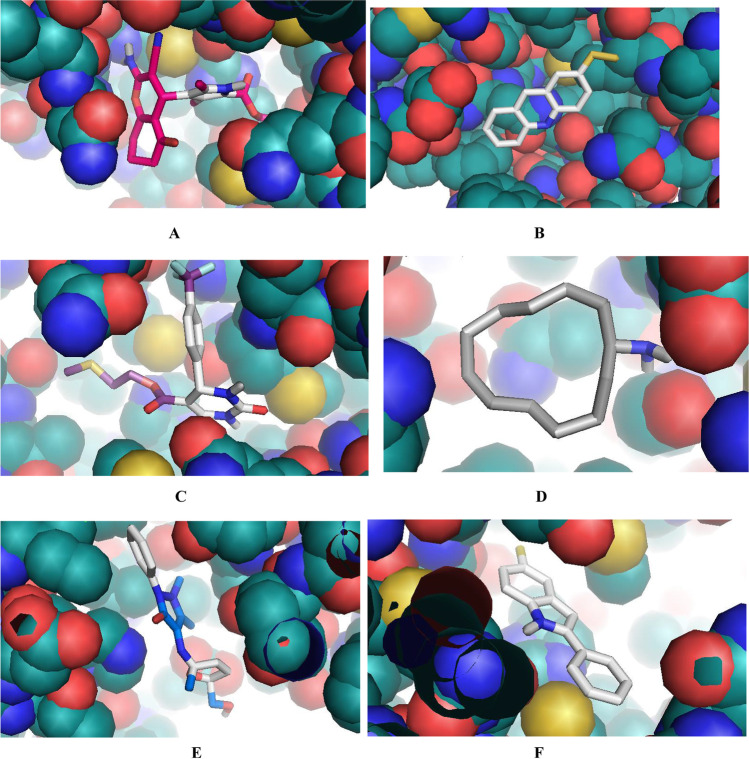
The molecular docking analysis and visualization of the selected proteins of Coronavirus-2 with the phytochemicals identified from the six medicinal plants (Azadirachta indica, Corchorus olitorius, Cassia alata, Phyllanthus amarus, Vernonia amygdalina, and Anacardium occidentale) were showed in Tables 7, 8, 9, 10, 11 and 12. The results revealed that the phytochemicals demonstrated high binding affinities against the proteins. This indicates that these phytochemicals could have substantial inhibitory effects against these proteins thus mitigating their pathogenic properties, ultimately leading to suppression of the pathogenesis of COVID-19.
Table 7.
Molecular docking score of the phytochemicals of Azadirachta indica against proteins of Coronavirus-2
| Phytochemicals | 2AJF | 6LU7 | 6VXS | 7BTF |
|---|---|---|---|---|
| Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | |
| (4-Methoxymethoxy-hex-5-ynylidene) -cyclohexane | − 5.8 | − 4.3 | − 5.1 | − 4.8 |
| 1,1-Di(isobutyl)acetone | − 5.5 | − 4.9 | − 5 | − 5.6 |
| 1,1-Diisobutoxy-isobutane | − 4.9 | − 4.8 | − 5 | − 5.2 |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | − 7 | − 6.4 | − 7.3 | − 7.8 |
| 1,4-Cyclohexanedimethanamine | − 4.9 | − 4.6 | − 4.7 | − 4.7 |
| 1-Butoxy-1-isobutoxy-butane | − 4.6 | − 4 | − 4.5 | − 4.5 |
| Ethane, 1-chloro-2-isocyanato- | − 3.4 | − 3.3 | − 3 | − 3.8 |
| 1-Methoxymethoxy-oct-2-yne | − 4.9 | − 3.9 | − 3.9 | − 4.6 |
| 1-Pentadecyne | − 4.2 | − 3.6 | − 4.1 | − 4.3 |
| Cyclohexanol, 1R-4-acetamido-2,3-cis-epoxy- | − 5.3 | − 5.1 | − 4.7 | − 4.8 |
| 1RA23A_Cyclohexanol | − 5.5 | − 5.2 | − 4.8 | − 5.1 |
| Butane, 2,2′-thiobis- | − 3.8 | − 3.4 | − 4 | − 4 |
| 2-Butynamide, N-methyl- | − 4.5 | − 3.5 | − 3.8 | − 4.3 |
| 1-Heptene, 2-methyl- | − 5 | − 3.6 | − 3.9 | − 4.5 |
| 3-Hexanone, 2-methyl- | − 4.4 | − 3.8 | − 3.9 | − 4.6 |
| 2_methylphenylester | − 5.9 | − 6.1 | − 6.2 | − 6.4 |
| 2-Nonanone | − 4.4 | − 3.8 | − 4.1 | − 4.7 |
| 2-Propen-1-amine | − 3.4 | − 3.3 | − 2.9 | − 3.5 |
| 2-Propenal | − 3 | − 2.6 | − 2.4 | − 3.2 |
| 3,4-Dimethylpent-2-en-1-ol | − 4.5 | − 4.4 | − 4.2 | − 4.7 |
| 3_Hydroxy_3_trifluoromethyl_2_oxa_ spiro[5_5]undecan_5_one | − 7.1 | − 6.3 | − 6.5 | − 6.9 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | − 5.8 | − 5.3 | − 5.2 | − 6.1 |
| 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl ester_yl)_3_5_dimethyl_1H_pyrrole_2_carboxylic acid ethyl ester | − 6.8 | − 7.6 | − 7.9 | − 7.5 |
| 4a-Methyl-6,8-dioxa-3-thia-bicyclo(3,2,1)octane | − 4.1 | − 4 | − 3.6 | − 4.1 |
| 5_Methoxymethoxyhexa_2_3_diene | − 4.1 | − 3.4 | − 3.7 | − 4.2 |
| 9,12-Octadecadienoic acid (Z,Z)- | − 7.3 | − 5.9 | − 7.3 | − 6.4 |
| beta.-D-Ribopyranoside, methyl 2, 3,4-tri-O-methyl- | − 4.2 | − 4.1 | − 3.8 | − 4.2 |
| Butane, 1,1-dibutoxy- | − 4.3 | − 3.9 | − 4.3 | − 4.2 |
| Chloroacetic acid | − 3 | − 3.1 | − 3 | − 3.9 |
| Chloroacetic acid, 1-cyclopentylet hyl ester | − 5.1 | − 4.4 | − 4.8 | − 5.2 |
| Chloromethyl octyl ether | − 4.6 | − 3.7 | − 4.1 | − 4.5 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | − 5.7 | − 4.9 | − 5.4 | − 5.1 |
| Cyclohexanol_ 2_(2_propynyloxy)__ trans_ | − 5.2 | − 4.4 | − 5.1 | − 4.7 |
| Cyclohexanone, 2-(2-propenyl)- | − 5.3 | − 4.6 | − 4.7 | − 6 |
| Dodecanoic acid | − 6.4 | − 4.8 | − 5.6 | − 6 |
| Ethyl Acetate | − 3.7 | − 3.3 | − 2.8 | − 3.8 |
| Ethylbenzene | − 6.1 | − 4.4 | − 4.5 | − 5.5 |
| Furo[′,3′:4,5]thiazolo[3,2-g]purine-8-methanol, 4-amino-6.alpha.,7,8,9a-tetrahydro-7-hydroxy-, [6aS(6a.alpha.,7.alpha.,8.beta.,9a.alpha.)]- | − 6 | − 6.1 | − 5.4 | − 6.4 |
| Hexadecanoic acid, 3,7,11,15-tetramethyl-, methyl ester | − 6.5 | − 5.5 | − 5.8 | − 6.1 |
| Methyl 6_O_[1_methylpropyl]__beta_ _d_galactopyranoside | − 5.6 | − 5.7 | − 5.6 | − 5.7 |
| n-Butyl ether | − 4 | − 3.6 | − 3.8 | − 3.7 |
| n-Decanoic acid | − 5.9 | − 5 | − 5.5 | − 5.4 |
| N1,N1-Dimethyl-N2-(1-phenyl–ethyl) -ethane-1,2-diamine | − 6.3 | − 5.6 | − 5.5 | − 5.7 |
| Nonanoic acid | − 5.5 | − 4.8 | − 5.2 | − 5 |
| o-Xylene | − 6.4 | − 4.6 | − 4.5 | − 5.7 |
| Propanoic acid | − 3.6 | − 3.2 | − 3.1 | − 4.1 |
| Thioctic acid | − 5.3 | − 4.6 | − 4.9 | − 5 |
| Undec-10-ynoic acid | − 5.6 | − 4.9 | − 5.5 | − 5.5 |
Table 8.
Molecular docking analysis of the phytochemicals of Corchorus olitorius against proteins of Coronavirus-2
| Phytochemicals | 2AJF | 6LU7 | 6VXS | 7BTF |
|---|---|---|---|---|
| Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | |
| 1,1-Cyclohexanedicarbonitrile | − 5.1 | − 5.1 | − 4.6 | − 5.4 |
| 1-Butoxy-1-isobutoxy-butane | − 4.8 | − 4.6 | − 4.7 | − 4.8 |
| Ethane, 1-chloro-2-isocyanato- | − 3.6 | − 3.3 | − 3 | − 3.9 |
| Tridecyl (E)-2-methylbut-2-enoate | − 3.6 | − 3.1 | − 3.2 | − 3.5 |
| 11-Bromoundecanoic acid | − 6 | − 4.9 | − 5.4 | − 5.6 |
| 12, 12-Dimethoxydodecanoic acid, methyl ester | − 5.1 | − 4 | − 4.5 | − 4.4 |
| 2-Ethylacridine | − 7 | − 6.4 | − 6.9 | − 7.1 |
| 3-Hexanone, 2-methyl- | − 4.7 | − 3.8 | − 3.9 | − 4.5 |
| 2-Propen-1-amine | − 3.1 | − 3.3 | − 2.9 | − 3.1 |
| 2-Propenal | − 2.9 | − 2.6 | − 2.4 | − 3.2 |
| 3_4_dihydroxy_1_Butyl | − 3.8 | − 4 | − 3.4 | − 4.1 |
| 3-Phenylpropionic acid, 4-methoxy- 2-methylbutyl ester | − 5.7 | − 4.9 | − 5.7 | − 6.1 |
| 4-Ethyl-4-methyl-5-methylene-[1,3] dioxolan-2-one | − 4.8 | − 4.4 | − 4.1 | − 5 |
| 9-Bromononanoic acid | − 5.5 | − 4.4 | − 5.3 | − 5.9 |
| Acetamide, N-(4-hydroxycyclohexyl) -, cis- | − 5.6 | − 4.8 | − 5 | − 4.9 |
| Acetamide, N-cyclohexyl- | − 4.9 | − 4.3 | − 5 | − 5.1 |
| Acetamide, N-cyclohexyl-2-[(2-furanylmethyl)thio]- | − 5.9 | − 5.2 | − 5.5 | − 5.8 |
| Acetic acid_ butyl ester | − 3.8 | − 3.4 | − 3.5 | − 4.2 |
| Acetic acid_ pentyl ester | − 4.1 | − 3.6 | − 3.9 | − 4.6 |
| Benzene, 1,3-dimethyl- | − 6.2 | − 4.7 | − 4.8 | − 5.6 |
| Butane, 1, 1-dibutoxy- | − 4.7 | − 4 | − 4.2 | − 4 |
| Butyric acid, 2,2-dimethyl-vinyl ester | − 4.2 | − 4.2 | − 4 | − 4.8 |
| ethyl Acetate | − 3.1 | − 2.9 | − 2.8 | − 3.7 |
| Formic acid, 3,3-dimethylbut-2-yl ester | − 4 | − 3.9 | − 3.8 | − 4.3 |
| Hexadecane, 1, 1- dimethoxy- | − 4.8 | − 3.8 | − 4.5 | − 4.5 |
| Isopropyl isothiocyanate | − 3.3 | − 3.4 | − 3.1 | − 3.6 |
| L-Lyxose | − 4.3 | − 4.9 | − 4 | |
| n-Butyl ether | − 3.8 | − 3.6 | − 3.9 | − 3.6 |
| N-Cyclododecylacetamide | − 6.3 | − 6 | − 5.8 | − 6.3 |
| o-Xylene | − 5.2 | − 4.6 | − 4.8 | − 5.6 |
| Octadecanoic acid | − 7 | − 5.7 | − 4.5 | − 6.9 |
| pyrrolidine_ 2_5_dione | − 4.4 | − 4 | − 5.9 | − 4.7 |
| Tridecyl (E)_2_methylbut_2_enoate | − 4.5 | − 3.9 | − 3.9 | − 4.8 |
Table 9.
Molecular docking analysis of the phytochemicals of Cassia alata against proteins of Coronavirus-2
| Phytochemicals | 2AJF | 6LU7 | 6VXS | 7BTF |
|---|---|---|---|---|
| Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | − 6.9 | − 6.3 | − 7.2 | − 7.8 |
| 1-Butoxy-1-isobutoxy-butane | − 4.6 | − 4.2 | − 4.5 | − 4.4 |
| 1-Oxa-4-azaspiro[4.5]decan-4-oxyl, 3,3-dimethyl-8-oxo- | − 5.5 | − 5.2 | − 5.1 | − 5.6 |
| 2-Octanol, 8,8-dimethoxy-2,6-dimethyl- | − 5.4 | − 4.9 | − 5.2 | − 5.4 |
| 3-Hexanone, 2-methyl- | − 4.4 | − 3.8 | − 3.9 | − 4.5 |
| 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2_methylsulfanyl_ethyl ester | − 7.1 | − 6.9 | − 7.5 | − 7.2 |
| Acetaldehyde, butylhydrazone | − 4.3 | − 3.6 | − 3.8 | − 4.5 |
| Acetic acid, butyl ester | − 4.2 | − 3.4 | − 3.6 | − 4.2 |
| Butane, 1, 1-dibutoxy- | − 4.5 | − 3.9 | − 4.3 | − 4.3 |
| Butyric acid, 2, 2-dimethyl-, vinyl ester | − 4.2 | − 4.2 | − 4 | − 4.4 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | − 5.6 | − 4.9 | − 5.4 | − 5.8 |
| Cyclohexaneacetic acid, butyl ester | − 5.2 | − 4.1 | − 5.2 | − 5.4 |
| Cyclopentaneundecanoic acid | − 7.6 | − 5.6 | − 6.6 | − 6.3 |
| D-Arabinitol | − 4.4 | − 5.2 | − 4 | − 4.5 |
| Ethanol, 2-phenoxy-, propanoate | − 5.1 | − 4.5 | − 5.2 | − 5.6 |
|
Furo[2′,3′:4,5]thiazolo[3,2-g]purine-8-methanol, 4-amino-6.alpha.,7,8,9a-tetrahydro-7-hydroxy-, [6aS-(6a.alpha.,7.alpha.,8.beta.,9a.alph a.)]- |
− 5.9 | − 6.1 | − 5.4 | − 6.4 |
| Hexyl 2-hydroxyethyl sulfide | − 4.6 | − 4.5 | − 4.4 | − 4.5 |
| n-Butyl ether | − 4.1 | − 3.5 | − 3.9 | − 3.6 |
| o-Xylene | − 5.3 | − 4.6 | − 4.5 | − 5.7 |
| Paramethadione | − 4.7 | − 4.5 | − 4.3 | − 4.9 |
| Propanoic acid, 2-methylpropyl ester | − 4.2 | − 3.7 | − 4 | − 4.6 |
| Pyrazol-3(2H)-one, 4-(2-furfurylidenamino)-1,5-dimethyl-2-phenyl- | − 6.4 | − 6.9 | − 6.8 | − 6.5 |
| Ribitol | − 5 | − 5.2 | − 4.8 | − 5 |
| Tridecyl (E)-2-methylbut-2-enoate | − 5.1 | − 4.2 | − 4.7 | − 5.4 |
Table 10.
Molecular docking analysis of the phytochemicals of Phyllanthus amarus against proteins of Coronavirus-2
| Phytochemicals | 2AJF | 6LU7 | 6VXS | 7BTF |
|---|---|---|---|---|
| Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | |
| 1,3-Dioxane, 2-ethyl-5-methyl- | − 4.2 | − 3.9 | − 4 | − 4.1 |
| 1, 3-Hexanediol, 2-ethyl- | − 4.8 | − 4.7 | − 4.5 | − 4.6 |
| 1,6-Anhydro-2,3-dideoxy-.beta.-D-erythro-hexopyranose | − 4.9 | − 4.5 | − 4.1 | − 4.8 |
| 1-Butoxy-1-isobutoxy_-butane | − 4.6 | − 4.4 | − 4.7 | − 4.6 |
| 1-Hexene, 3, 4-dimethyl- | − 4.7 | − 4.2 | − 4.1 | − 5 |
| 1-Undecene, 2-methyl- | − 5 | − 3.9 | − 4.5 | − 4.3 |
| 2,4-Dipropyl-5-ethyl-1,3-dioxane | − 5 | − 4.6 | − 4.7 | − 4.8 |
| 2_Methylvaleroyl chloride | − 4.3 | − 4.1 | − 3.8 | − 4.4 |
| 2H-Pyran-2-one, tetrahydro-5,6-dimethyl-, trans- | − 4.9 | − 4.2 | − 4.2 | − 5.4 |
| 3-Hexanone, 2-methyl- | − 4.4 | − 3.8 | − 3.9 | − 4.5 |
| 6-Bromohexanoic acid, hexyl ester | − 5 | − 3.8 | − 4.7 | − 4.6 |
| Acetic acid, butyl ester | − 3.7 | − 3.3 | − 3.5 | − 4.2 |
| Butane, 1,1′-[ethylidenebis(oxy)]bis- | − 4.2 | − 3.7 | − 4.2 | − 4.3 |
| Butane, 1,1-dibutoxy- | − 4.5 | − 3.8 | − 4.1 | − 4.1 |
| Butanoic acid, butyl ester | − 4 | − 3.7 | − 4.1 | − 4.2 |
| Butyric acid, 2,2-dimethyl-, vinylester | − 4.4 | − 4.2 | − 4 | − 4.5 |
| Cyclododecylamine | − 6 | − 5.2 | − 5.5 | − 6 |
| Decanoic acid, propyl ester | − 4.8 | − 4.1 | − 4.2 | − 4.4 |
| Formic acid, 3,3-dimethylbut-2-ylester | − 4.1 | − 3.9 | − 3.8 | − 4.1 |
| n-Butyl ether | − 4 | − 3.6 | − 3.7 | − 3.7 |
| n-Butyl isobutyl sulfide | − 3.8 | − 3.5 | − 4 | − 3.9 |
| Neopentyl isothiocyanate | − 4.3 | − 3.8 | − 3.7 | − 3.8 |
| o-Xylene | − 6.4 | − 4.6 | − 4.5 | − 5.6 |
| Pentane, 1-bromo-3,4-dimethyl- | − 4.3 | − 4.1 | − 4 | − 4.7 |
| Pentane, 2,3-dimethyl- | − 4.4 | − 3.7 | − 3.8 | − 4.8 |
| Propane, 1-isothiocyanato- | − 3.4 | − 3.2 | − 3.2 | − 3.3 |
Table 11.
Molecular docking analysis of the phytochemicals of Vernonia amygdalina against proteins of Coronavirus-2
| Phytochemicals | 2AJF | 6LU7 | 6VXS | 7BTF |
|---|---|---|---|---|
| Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | |
| (4-Methoxymethoxy-hex-5-ynylidene)-cyclohexane | − 5.5 | − 4.6 | − 5.4 | − 5.3 |
| .beta.-D-Xylopyranoside, methyl 2,3,4-tri-O-methyl- | − 3.8 | − 4 | − 3.8 | − 4.5 |
| 1,4-Cyclohexanedimethanamine | − 4.9 | − 4.7 | − 4.7 | − 4.6 |
| 1,6-Anhydro-2,3-dideoxy-.beta.-D-erythro-hexopyranose | − 4.9 | − 4.5 | − 4.1 | − 4.7 |
| 1-Butoxy-1-isobutoxy-butane | − 4.3 | − 3.9 | − 4.7 | − 4.6 |
| 1-Butyl-3,4-dihydroxy-pyrrolidine- 2,5-dione | − 5.3 | − 5.1 | − 4.8 | − 6.2 |
| 1-Hexene, 3,4-dimethyl- | − 4.7 | − 4.2 | − 4.1 | − 5 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | − 5.3 | − 4.9 | − 5 | − 5.7 |
| 1_Propanol_ 2_2_dimethyl__ acetate | − 4.1 | − 3.8 | − 3.8 | − 4.3 |
| 2-Butenoic acid, butyl ester | − 4.3 | − 3.7 | − 3.9 | − 4 |
| 2-Butynamide, N-methyl- | − 4.1 | − 3.5 | − 3.8 | − 4.7 |
| 2-Propanone, propylhydrazone | − 4.2 | − 3.7 | − 4.1 | − 4.2 |
| 2-Propen-1-amine | − 3.1 | − 3 | − 2.9 | − 3.4 |
| 2-Propenal | − 3 | − 2.6 | − 2.4 | − 3.2 |
| 2-Tetradecanol | − 6.4 | − 5.1 | − 5.9 | − 6.1 |
| 2H-1,2,3-Triazol-4-amine, 2-cyclohexyl-5-nitro-, 1-oxide | − 6.4 | − 5.6 | − 5.9 | − 6.6 |
| 3,4,5-Trimethyldihydrofuran-2-one | − 4.5 | − 4.2 | − 4.1 | − 5.1 |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | − 4.7 | − 4.5 | − 4.4 | − 5.1 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | − 5.8 | − 5.3 | − 5.2 | − 5.9 |
| 8-Chlorocapric acid | − 5.7 | − 5.1 | − 5.3 | − 5.3 |
| Acetamide, N-cyclohexyl- | − 5.3 | − 4.3 | − 5 | − 5 |
| Acetamide, N-cyclohexyl-2-[(2-furanylmethyl)thio]- | − 5.9 | − 5.6 | − 5.4 | − 6 |
| Acetic acid, butyl ester | − 3.7 | − 3.2 | − 3.5 | − 4 |
| Aziridine, 2-methyl- | − 3.3 | − 2.6 | − 2.7 | − 3.3 |
| Butane, 1,1-dibutoxy- | − 4.2 | − 3.7 | − 4.1 | − 4.1 |
|
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
− 6.1 | − 6.2 | − 5.7 | − 6.6 |
| Cyclobutanone, 2-methyl-2-oxiranyl | − 4.3 | − 4 | − 3.9 | − 4.5 |
| Cyclohexane acetic acid, butyleste | − 5.4 | − 4.4 | − 5.2 | − 4.8 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | − 5.2 | − 4.9 | − 5.4 | − 5.1 |
| Cyclohexaneacetic acid_ butyl ester | − 5.6 | − 4.6 | − 5.2 | − 5.1 |
| Cyclohexanone, 2-(1-mercapto-1-methylethyl)-5-methyl-, trans- | − 5.2 | − 5.3 | − 5.1 | − 5.7 |
| Cyclohexanone, 2-(2-propenyl)- | − 5.1 | − 4.5 | − 4.7 | − 5.4 |
| Cyclopentane, 1-methyl-2-(4-methylpentyl)-, trans- | − 5.4 | − 4.8 | − 5.1 | − 5.5 |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | − 5.2 | − 4.8 | − 5.1 | − 5.3 |
| Cyclopentaneundecanoic acid | − 7.7 | − 5.9 | − 6.8 | − 6.3 |
| Dodecanoic acid | − 6.5 | − 5.1 | − 5.4 | − 5.3 |
| Ethanamine, N-pentylidene- | − 3.9 | − 3.3 | − 3.8 | − 3.6 |
| Ethane, 1-chloro-2-isocyanato- | − 3.5 | − 3.3 | − 3 | − 3.8 |
| Ethanol, 2-bromo- | − 2.9 | − 2.9 | − 2.7 | − 3.3 |
| Ethyl Acetate | − 3.5 | − 3 | − 2.9 | − 3.9 |
| Ethylbenzene | − 6.1 | − 4.4 | − 4.5 | − 5.5 |
|
Formic acid, 3,3-dimethylbut-2-yl ester |
− 4.1 | − 3.9 | − 3.8 | − 4.2 |
| Morpholine | − 3.2 | − 3.3 | − 3 | − 4.2 |
| n-Butyl ether | − 4.1 | − 3 | − 3.7 | − 4.5 |
| N-Cyclododecylacetamide | − 6.4 | − 6 | − 5.8 | − 6.4 |
| n-Decanoic acid | − 6.1 | − 4.9 | − 5.6 | − 5.9 |
| Oxirane, (1-methylethyl)- | − 3.5 | − 3.2 | − 4.8 | − 3.7 |
| Oxonin, 4,5,6,7-tetrahydro-, (Z,Z)32 | − 4.9 | − 4.6 | − 3.3 | − 5.4 |
| Pentadecanoic acid | − 6.8 | − 5.3 | − 4.3 | − 6.2 |
| Pentane, 1-bromo-3,4-dimethyl- | − 4.3 | − 4.1 | − 5.9 | − 4.7 |
| Propane, 1,2-dichloro-2-fluoro- | − 3.7 | − 3.6 | − 4 | − 3.7 |
| Pyrazol-3(2H)-one, 4-(5-hydroxymethylfurfur-2-ylidenamino)-1,5-dimethyl-2-phenyl- | − 6.7 | − 6.7 | − 3.6 | − 6.3 |
| Succinic acid, hexyl 3-oxobut-2-yl-ester | − 4.7 | − 4.1 | − 6.7 | − 5.5 |
| Thiazole, 2-ethyl-4,5-dihydro- | − 3.7 | − 3.2 | − 4.8 | − 3.7 |
| Tridecyl (E)-2-methylbut-2-enoate | − 5.5 | − 4.3 | − 3.3 | − 5.1 |
Table 12.
Molecular docking analysis of the phytochemicals of Anacardium occidentale against proteins of Coronavirus-2
| Phytochemicals | 2AJF | 6LU7 | 6VXS | 7BTF |
|---|---|---|---|---|
| Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | Binding energies (kcal/mol) | |
| .alpha.-D-Glucopyranose, 4-O-.beta.-D-galactopyranosyl | − 6.3 | − 6.4 | − 6.3 | − 6.2 |
| 1-Benzenesulfonyl-1H-pyrrole | − 6.7 | − 5.6 | − 5.6 | − 6 |
| 1-Butoxy-1-isobutoxy-butane | − 4.6 | − 4.1 | − 4.6 | − 4.4 |
| 1-Methoxy-3-(2-hydroxyethyl)nonane | − 5.7 | − 4.8 | − 5 | − 5.6 |
| 1-Octene, 2-methyl- | − 4.2 | − 3.7 | − 4.1 | − 4.6 |
| 1-Oxa-4-azaspiro[4.5]decan-4-oxyl, 3,3-dimethyl-8-oxo- | − 5.1 | − 5.3 | − 5.2 | − 6 |
| 1-Pentadecene, 2-methyl- | − 5.6 | − 3.8 | − 4.6 | − 4.8 |
| 1-Pentene, 5-chloro-4-(chloromethyl)-2,4-dimethyl- | − 4.9 | − 4 | − 4.2 | − 4.7 |
| 1-Phenyloxycarbonyl-7-pentyl-7-aza bicyclo[4.1.0]heptane | − 6.4 | − 6 | − 6.7 | − 6.2 |
| 1-Propanol,2,2-dimethyl- acetate | − 4 | − 3.9 | − 3.7 | − 4.3 |
| 1H-Imidazole, 2-ethyl-4,5-dihydro- | − 4.3 | − 3.6 | − 3.5 | − 4.6 |
| 1H-Indole, 5-methyl-2-phenyl- | − 6.9 | − 7.2 | − 7.4 | − 7.1 |
|
2(1H)-Naphthalenone, octahydro-4a- methyl-7-(1-methylethyl)-, (4a.alp ha.,7.beta.,8a.beta.) - |
− 6.2 | − 6 | − 6.2 | − 6.7 |
| 2-Butylthiazoline | − 4.1 | − 3.7 | − 3.7 | − 4 |
| 2-Butynamide, N-methyl- | − 4.1 | − 3.5 | − 3.8 | − 4.3 |
| 2-Propanone, propylhydrazone | − 4.2 | − 3.6 | − 4.1 | − 4.3 |
| 2-Propen-1-amine | − 3.4 | − 3 | − 2.9 | − 3.5 |
| 3-Oxatricyclo[4.2.0.0(2,4)]octan-7-one | − 4.7 | − 4.5 | − 4.4 | − 5.1 |
| 3H-Naphth[1,8a-b]oxiren-2(1aH)-one, hexahydro- | − 5.8 | − 5.3 | − 5.3 | − 6.1 |
| 4.beta.,5-Dimethyl-6,8-dioxa-3-thiabicyclo(3,2,1)octane 3-oxide | − 4.7 | − 4.6 | − 4.1 | − 4.7 |
| 4-t-Butylcyclohexylamine | − 5.6 | − 5.1 | − 5.3 | − 5.7 |
| Acetaldehyde, butylhydrazone | − 4.4 | − 3.8 | − 3.8 | − 4.2 |
| Acetamide, N-(4-hydroxycyclohexyl) -, cis- | − 5.6 | − 4.9 | − 5 | − 5.1 |
| Acetic acid, butyl ester | − 3.9 | − 3.4 | − 3.5 | − 3.8 |
| Butane, 1,1-dibutoxy- | − 4.2 | − 3.7 | − 4.3 | − 4 |
| Butane, 2,2′-thiobis- | − 4 | − 3.5 | − 3.8 | − 4.2 |
| Chloroacetic acid, 1-cyclopentylethyl ester | − 5.2 | − 4.5 | − 4.7 | − 5.2 |
| Cyclobutanone, 2-methyl-2-oxiranyl | − 4.3 | − 4.1 | − 3.9 | − 4.5 |
| Cyclododecanol, 1-aminomethyl- | − 6.1 | − 5.8 | − 6.1 | − 6.8 |
| Cyclohexane, 1R-acetamido-2,3-cis-epoxy-4-cis-formyloxy- | − 5.8 | − 4.9 | − 5.4 | − 5.3 |
| Cyclohexanone, 2-(2-propenyl)- | − 4.9 | − 4.4 | − 4.7 | − 5.9 |
| Cyclopentaneethanol, 2-(hydroxymethyl)-.beta.,3-dimethyl- | − 5.2 | − 4.8 | − 5.1 | − 5.2 |
| Cyclopentaneundecanoic acid | − 7.3 | − 5.7 | − 6.6 | − 7.2 |
| Dimethylamine, N-(neopentyloxy)- | − 3.8 | − 3.7 | − 3.6 | − 4.2 |
| Dodecanoic acid, 1,2, 3-propanetriyl ester | − 5.5 | − 3.9 | − 4.5 | − 5 |
| Ethanamine, N-pentylidene- | − 3.8 | -3.3 | − 3.8 | − 4.6 |
| Ethane, 1-chloro-2-isocyanato- | − 3.4 | − 3 | − 3 | − 3.8 |
| Ethanol, 2-butoxy- | − 4.2 | − 3.6 | − 3.8 | − 4.1 |
| Ethyl Acetate | 3.3 | − 3.2 | − 2.8 | − 3.7 |
| Ethylbenzene | − 6.1 | − 4.4 | − 4.5 | − 5.5 |
| Formic acid, 3,3-dimethylbut-2-yl ester | − 3.9 | − 3.8 | − 3.8 | − 4.2 |
| n-Butyl ether | − 4 | − 3.4 | − 3.8 | − 3.8 |
| n-decanoic acid | − 5.8 | − 4.9 | − 5.6 | − 6 |
| Neopentyl isothiocyanate | − 3.8 | − 3.6 | − 3.7 | − 3.9 |
| Nonanoic acid | − 5.5 | − 4.8 | − 5 | − 5.2 |
| o-Xylene | − 5.2 | − 4.6 | − 4.5 | − 5.7 |
| p-Xylene | − 6.1 | − 4.5 | − 4.5 | − 5.6 |
| Pyrazol-3(2H)-one, 4-(5-hydroxymethylfurfur-2-ylidenamino)-1,5-dimethyl-2-phenyl- | − 6.7 | − 6.7 | − 6.7 | − 6.6 |
| Pyrido[2,3-d]pyrimidine, 4-phenyl- | − 6.8 | − 6.3 | − 6.6 | − 7 |
| Spiro[2.3]hexan-4-one, 5,5-diethyl | − 4.6 | − 4.1 | − 4.5 | − 4.9 |
| Succinic acid,butyl decyl ester | − 5.4 | − 4.2 | − 4.7 | − 4.8 |
| Thiazole, 2-ethyl-4,5-dihydro- | − 3.4 | − 3.2 | − 3.3 | − 3.5 |
| Tridecanoic acid | − 6.5 | − 5.1 | − 6 | − 6 |
| Z,E-7,11-Hexadecadien-1-yl acetate | − 5.7 | − 4.5 | − 5 | − 5.2 |
Molecular docking analysis of SARS coronavirus spike receptor-binding domain (2AJF)
The in silico analysis of 2AJF binding with the best ligands with the highest docking scores is shown in Fig. 13, Fig. 14, and Table 13. Out of the investigated phytochemicals from Azadirachta indica, 9,12-Octadecadienoic acid (Z,Z)- exhibited the best docked score (− 7.3 kcal/mol) with the SARS coronavirus spike receptor-binding domain (2AJF). The amino acid residues- ALA-348, SER-44, PHE-40, PHE-390, ASN-394, TRP-349, GLU-37, GLY-352, ASP-350, LEU-351, and ARG-393 participated extensively in the interaction at the binding pocket of 2AJF. N-Cyclododecylacetamide from Corchorus olitorius displayed the best docked score (− 6.4 kcal/mol) with 2AJF having TRP-69, PHE-40, ASP-350, LEU-73, PHE-390, ARG-393, LEU-391, LEU-100, ALA-99, SER-77, and PHE-32 amino acid residues participating in the interaction. Similarly, cyclopentaneundecanoic acid from Cassia alata demonstrated the best docked score (− 7.6 kcal/mol) with 2AJF having ASP-350, PHE-30, TRP-69, ARG-393, PHE-390, LEU-391, LEU-73, ALA-99, GLN-102, GLN-101, SER-77, and LEU-100 amino acid residues participating in the interaction. Of all the phytochemicals present in Phyllanthus amarus, o-Xylene revealed the best docked score (− 6.4 kcal/mol) with 2AJF. PRO-415, THR-414, ALA-413, THR-434, GLU-345, PHE-438, and HIS-540 amino acid residues participating in the interaction at the binding pocket of 2AJF. Cyclopentaneundecanoic acid from Vernonia amygdalina displayed the best docked score (− 7.7 kcal/mol) with 2AJF having PHE-69, PHE-40, PHE-32, LEU-73, SER-77, LEU-100, ALA-99, GLN-102, ASN-394, ARG-393, ASP-350, PHE-390, and LEU-391 amino acid residues participating in the interaction. Furthermore, cyclopentaneundecanoic acid from Anacardium occidentale demonstrated the best docked score (− 7.3 kcal/mol) with 2AJF having LEU-351, PHE-40, TRP-69, ASP-350, GLU-37, GLY-352, PHE-390, ARG-393, LEU-391, LEU-73, LEU-100, and ALA-99 amino acid residues participating in the interaction.
Fig. 13.
Docking analysis and visualization of 2AJF binding with the best ligands with the highest docking score
Fig. 14.
Binding-interaction analysis of 2AJF binding with the best ligands with the highest docking score
Table 13.
Binding analysis of the best ligand with 2AJF (SARS coronavirus spike receptor-binding domain)
| ID | Ligand | Amino acid residues involved in the interaction | Polar information |
|---|---|---|---|
| A | 9,12-Octadecadienoic acid (Z,Z)- | ALA-348, SER-44, PHE-40, PHE-390, ASN-394, TRP-349, GLU-37, GLY-352, ASP-350, LEU-351, ARG-393 |
ASP-350 2.8 Å, ARG-393 3.5 Å |
| B | N-Cyclododecylacetamide | TRP-69, PHE-40, ASP-350, LEU-73, PHE-390, ARG-393, LEU-391, LEU-100, ALA-99, SER-77, PHE-32 | ASP-350 2.4 Å |
| C | Cyclopentaneundecanoic acid | ASP-350, PHE-30, TRP-69, ARG-393, PHE-390, LEU-391, LEU-73, ALA-99, GLN-102, GLN-101, SER-77, LEU-100 |
GLN-101 4.6 Å ALA-99 2.4 Å LEU-100 2.3 Å |
| D | o-Xylene | PRO-415, THR-414, ALA-413, THR-434, GLU-345, PHE-438, HIS-540 | No polar contacts |
| E | Cyclopentaneundecanoic acid | PHE-69, PHE-40, PHE-32, LEU-73, SER-77, LEU-100, ALA-99, GLN-102, ASN-394, ARG-393, ASP-350, PHE-390, LEU-391 | No polar contacts |
| F | Cyclopentaneundecanoic acid | LEU-351, PHE-40, TRP-69, ASP-350, GLU-37, GLY-352, PHE-390, ARG-393, LEU-391, LEU-73, LEU-100, ALA-99 |
ASP-350 2.8 Å ARG-393 3.5 Å |
Molecular docking analysis of SARS-COV2 major protease (6LU7)
Figure 15, Fig. 16, and Table 14 present the molecular docking analysis and visualization of 6LU7 binding with the best ligands with the highest docking scores. Of all the examined phytocompounds from Azadirachta indica, 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester displayed the highest docked score (− 7.6 kcal/mol) with the SARS-COV2 major protease (6LU7). MET-49, TYR-54, GLN-189, ARG-188, ASP-187, HIS-164, MET-165, GLU-166, HIS-163, LEU-141, SER-144, LYS-145, ASN-142, GLY-143, and CYS-145 amino acid residues participating in the interaction at the binding pocket of 6LU7. 2-Ethylacridine from Corchorus olitorius presented the highest affinity score (− 6.9 kcal/mol) with 6LU7 having ARG-105, ILE-106, GLN-110, OHE-294, ASP-153, ILE-152, ASN-151, and PHE-8 amino acid residues participating in the interaction. Equally, 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester from Cassia alata produced the greatest docked score (− 6.9 kcal/mol) with 6LU7 having PHE-140, LEU-141, HIS-172, GLU-166, MET-165, SER-144, ASN-142, GLY-143, CYS-145, HIS-163, HIS-164, GLN-189, HIS-41, THR-26, LEU-27, and MET-49 amino acid residues participating in the interaction. Out of all the phytocompounds present in Phyllanthus amarus, cyclododecylamine displayed the peak affinity score (− 5.2 kcal/mol) with 6LU7. PHE-294, GLN-110, ASN-151, THR-111, SER-158, LYS-102, and ASP-153 amino acid residues “participated” in the interaction at the binding pocket of 6LU7. N-Cyclododecylacetamide from Vernonia amygdalina exhibited the best docked score (− 6.0 kcal/mol) with 6LU7 having G;U-240, VAL-202, HIS-246, ILE-249, ILE-200, ASN-203, PRO-293, GLN-110, GLY-109, and PRO-108 amino acid residues participating in the interaction. Additionally, 1H-Indole, 5-methyl-2-phenyl- from Anacardium occidentale demonstrated the best docked score (− 7.2 kcal/mol) with 6LU7 having LEU-141, ASN-142, GLY-143, MET-165, HIS-164, CYS-145, HIS-41, GLN-189, TYR-54, ASP-187, ARG-188, and MET-49 amino acid residues participating in the interaction.
Fig. 15.
Docking analysis and visualization of 6LU7 binding with the best ligands with the highest docking score
Fig. 16.
Bind-interaction analysis of 6LU7 binding with the best ligands with the highest docking score
Table 14.
Binding analysis of the best ligand with 6lu7 (COVID-19 main protease)
| ID | S/N | Ligand | Amino acid residues involved in the interaction | Polar information |
|---|---|---|---|---|
| A | 1 | 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester |
MET-49, TYR-54, GLN-189, ARG-188, ASP-187, HIS-164, MET-165, GLU-166, HIS-163 LEU-141, SER-144, LYS-145, ASN-142, GLY-143, CYS-145 |
GLY-143 2.9 Å HIS-163 3.4 Å SER-144 2.7 Å |
| B | 2 | 2-Ethylacridine | ARG-105, ILE-106, GLN-110, OHE-294, ASP-153, ILE-152, ASN-151, PHE-8 | No polar contacts |
| C | 3 | 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester | PHE-140, LEU-141, HIS-172, GLU-166, MET-165, SER-144, ASN-142, GLY-143, CYS-145, HIS-163, HIS-164, GLN-189, HIS-41, THR-26, LEU-27, MET-49 | No polar contacts |
| D | 4 | Cyclododecylamine | PHE-294, GLN-110, ASN-151, THR-111, SER-158, LYS-102, ASP-153 |
ASP-153 2.5 Å SER-158 2.0 Å |
| E | 5 | N-Cyclododecylacetamide |
G;U-240, VAL-202, HIS-246, ILE-249, ILE-200, ASN-203, PRO-293, GLN-110, GLY-109 PRO-108 |
No polar contacts |
| F | 6 | 1H-Indole, 5-methyl-2-phenyl- | LEU-141, ASN-142, GLY-143, MET-165, HIS-164, CYS-145, HIS-41, GLN-189, TYR-54, ASP-187, ARG-188, MET-49 | No polar contacts |
Molecular docking analysis of ADP ribose phosphatase of NSP3 from SARS CoV-2 (6VXS)
The computational analysis of 6VXS binding with the best ligand with the highest docking score is shown in Fig. 17, Fig. 18, and Table 15. Out of the investigated phytochemicals from Azadirachta indica, 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl), 3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl ester exhibited the best docked score (− 7.9 kcal/mol) with the ADP ribose phosphatase of NSP3 from SARS CoV-2 (6VXS). The amino acid residues taking part in the interaction at the binding pocket of 6VXS are LEU-160, PHE-156, VAL-155, ALA-154, GLY-130, ILE-131, PHE-132, SER-128, ALA-38, VAL-49, ALA-129, LEU-160, PRO-136, and PRO-125. 2-Ethylacridine from Corchorus olitorius displayed the best docked score (− 7.5 kcal/mol) with 6VXS having LEU-160, PHE-156, VAL-155, ALA-154, ASP-22, ILE-23, and LEU-126 amino acid residues participating in the interaction. Similarly, 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester from Cassia alata demonstrated the best docked score (− 5.5 kcal/mol) with 6VXS having PRO-125, LEU-126, ILE-23, ALA-165, VAL-49, ALA-154, ALA-21, ASP-22, ASP-157, LEU-160, GLY-130, and VAL-155 amino acid residues participating in the interaction. Of all the phytochemicals present in Phyllanthus amarus, cyclododecylamine revealed the best docked score (− 5.5 kcal/mol) with 2AJF. VAL-49, PHE-156, VAL-155, ALA-154, ALA-21, ILE-23, and ASP-22 amino acid residues participated in the interaction at the binding pocket of 6VXS. Cyclopentaneundecanoic acid from Vernonia amygdalina displayed the best docked score (− 6.8 kcal/mol) with 6VXS having LEU-160, VAL-155, GLY-130, ILE-23, VAL-49, VAL-125, ALA-129, ALA-154, LEU-126, PHE-156, and ALA-21 amino acid residues participating in the interaction. Furthermore, 1H-Indole, 5-methyl-2-phenyl- from Anacardium occidentale demonstrated the best docked score (− 7.4 kcal/mol) with 6VXS having ALA-154, LEU-126, ILE-23, VAL-49, and LEU-160 amino acid residues participating in the interaction.
Fig. 17.
Docking analysis and visualization of 6VXS binding with the best ligands with the highest docking score
Fig. 18.
Bind-interaction analysis of 6VXS binding with the best ligands with the highest docking score
Table 15.
Binding analysis of the best ligand with 6VXS (ADP ribose phosphatase of NSP3 from SARS CoV-2)
| ID | S/N | Ligand | Amino acid residues involved in the interaction | Polar information |
|---|---|---|---|---|
| A | 1 | 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl), 3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl ester | LEU-160, PHE-156, VAL-155, ALA-154, GLY-130, ILE-131, PHE-132, SER-128, ALA-38, VAL-49, ALA-129, LEU-160, PRO-136, PRO-125 | ALA-129 3.5 Å |
| B | 2 | 2-Ethylacridine | LEU-160, PHE-156, VAL-155, ALA-154, ASP-22, ILE-23, LEU-126 | PHE-156 3.5 Å |
| C | 3 | 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester | PRO-125, LEU-126, ILE-23, ALA-165, VAL-49, ALA-154, ALA-21, ASP-22, ASP-157, LEU-160, GLY-130, VAL-155 | VAL-155 3.2 Å |
| D | 4 | Cyclododecylamine | VAL-49, PHE-156, VAL-155, ALA-154, ALA-21, ILE-23, ASP-22 | No polar contacts |
| E | 5 | Cyclopentaneundecanoic acid | LEU-160, VAL-155, GLY-130, ILE-23, VAL-49, VAL-125, ALA-129, ALA-154, LEU-126, PHE-156, ALA-21 | ALA-21 2.8 Å |
| F | 6 | 1H-Indole, 5-methyl-2-phenyl- | ALA-154, LEU-126, ILE-23, VAL-49, LEU-160 | ALA-154 2.5 Å |
Molecular docking analysis of SARS CoV-2 RNA-dependent RNA polymerase (7BTF)
Figure 19, Fig. 20, and Table 16 show the molecular docking analysis and visualization of 7BTF binding with the best ligand with the highest docking score. Of all the examined phytocompounds from Azadirachta indica, 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane displayed the highest docked score (− 7.8 kcal/mol) with the SARS CoV-2 RNA-dependent RNA polymerase (7BTF). The participating amino acid residues in the interaction at the binding pocket of 7BTF are ASN-710, LYS-38, ASN-36, LYS-47, PHE-45, VAL-39, PHE-45, PHE-32, THR-203, ASP-205, ASP-33, ILE-34, and LYS-70. 2-Ethylacridine from Corchorus olitorius presented the highest affinity score (− 7.1 kcal/mol) with 6LU7 having LEU-368, ALA-372, PHE-365, LEU-369, PHE-503, TYR-512, LEU-511, and TRP-506 amino acid residues participating in the interaction. Equally, 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester from Cassia alata produced the greatest docked score (− 7.2 kcal/mol) with 7BTF having LYS-70, LYS-47, ASP-205, THR-203, ASN-36, LYS-38, ASN-710, VAL-39, ASP-33, ILE-34, PHE-32, and PHE-45 amino acid residues participating in the interaction. Out of all the phytocompounds present in Phyllanthus amarus, cyclododecylamine displayed the peak affinity score (− 6.0 kcal/mol) with 7BTF. The amino acid residues- ALA-372, TRP-506, LEU-511, PHE-365, LEU-368, LEU-369, TYR-512, and PHE-503 participated extensively in the interaction at the binding pocket of 7BTF. Carbamic acid [1-(hydroxymethyl)-2-phenylethyl]-1,1-dimethylethyl ester, (s)- from Vernonia amygdalina exhibited the best docked score (− 6.6 kcal/mol) with 7BTF having PHE-45, LEU-46, PHE-32, ILE-34, PHE-45, LYS-47, THR-48, ASN-49, and LYS-70 amino acid residues participating in the interaction. Additionally, 1H-Indole, 5-methyl-2-phenyl- from Anacardium occidentale demonstrated the best docked score (− 7.1 kcal/mol) with 7BTF having ASP-215, THR-203, ASN-206, ASP-205, ILE-34, PHE-32, PHE-45, LYS-47, THR-48, and LYS-70 amino acid residues participating in the interaction.
Fig. 19.
Docking analysis and visualization of 7B binding with the best ligands with the highest docking score
Fig. 20.
Bind-interaction analysis of 7B binding with the best ligands with the highest docking score
Table 16.
Binding analysis of the best ligand with 7BTF (SARS CoV-2 RNA-dependent RNA polymerase)
| ID | S/N | Ligand | Amino acid residues involved in the interaction | Polar information |
|---|---|---|---|---|
| A | 1 | 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | ASN-710, LYS-38, ASN-36, LYS-47, PHE-45, VAL-39, PHE-45, PHE-32, THR-203, ASP-205, ASP-33, ILE-34, LYS-70 | ASN-36 2.1 Å |
| B | 2 | 2-Ethylacridine | LEU-368, ALA-372, PHE-365, LEU-369, PHE-503, TYR-512, LEU-511, TRP-506 | No polar contacts |
| C | 3 | 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester | LYS-70, LYS-47, ASP-205, THR-203, ASN-36, LYS-38, ASN-710, VAL-39, ASP-33, ILE-34, PHE-32, PHE-45 | ASN-36 2.1 Å |
| D | 4 | Cyclododecylamine | ALA-372, TRP-506, LEU-511, PHE-365, LEU-368, LEU-369, TYR-512, PHE-503 | No polar contacts |
| E | 5 |
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
PHE-45, LEU-46, PHE-32, ILE-34, PHE-45, LYS-47, THR-48, ASN-49, LYS-70 |
LEU-46 2.1 Å THR-48 2.3 Å |
| F | 6 | 1H-Indole, 5-methyl-2-phenyl- | ASP-215, THR-203, ASN-206, ASP-205, ILE-34, PHE-32, PHE-45, LYS-47, THR-48, LYS-70 |
ASP-215 2.7 Å THR-203 2.6 Å ASN-206 2.1 Å, 2.3 Å, 2.6 Å |
In silico toxicity and ADME of the phytochemicals with best docking score
Molecular properties of the phytochemicals with best docking score
Results in Table 17 showed the molecular properties of the phytochemicals with the best docking score. 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester was found to have the highest molecular weight of 374.364, followed by 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester with 365.394. Similarly, 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester and 2-Ethylacridine have the highest surface area of 151.006. Furthermore, 9,12-Octadecadienoic acid (Z,Z)- has the highest lipophilicity of 5.8845.
Table 17.
Molecular properties of phytochemicals with best docking scores
| Ligand | Molecular weight | Lipophilicity (Log P) | Number of rotatable bonds | Number of acceptors | Number of donors | Surface area |
|---|---|---|---|---|---|---|
| 9,12-Octadecadienoic acid (Z,Z)- | 280.452 | 5.8845 | 14 | 1 | 1 | 124.52 |
| N-Cyclododecylacetamide | 225.376 | 3.7958 | 1 | 1 | 1 | 100..189 |
| Cyclopentaneundecanoic acid | 254.414 | 5.1622 | 11 | 1 | 1 | 112.163 |
| o-Xylene | 106.166 | 2.30344 | 0 | 0 | 0 | 50.161 |
| 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester | 365.394 | 2.62302 | 3 | 6 | 2 | 151.006 |
| 2-Ethylacridine | 355.394 | 2.62302 | 3 | 6 | 2 | 151.006 |
|
6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester |
374.364 | 2.3295 | 5 | 4 | 2 | 146.541 |
| Cyclododecylamine | 163.339 | 3.6184 | 0 | 1 | 1 | 83.088 |
| 1H-Indole, 5-methyl-2-phenyl- | 207.275 | 4.14332 | 1 | 0 | 1 | 94.744 |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | 296.41 | 4.6911 | 3 | 2 | 0 | 132.629 |
|
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
251.326 | 2.1147 | 4 | 3 | 2 | 107..970 |
Predicted absorption properties of the phytochemicals with best docking score
The predicted absorption properties of the phytochemicals were reported in Table 18. The result showed that 9,12-Octadecadienoic acid (Z,Z)- has the highest water solubility value of − 5.862 while Cyclododecylamine has the lowest valve of − 2.651. O-Xylene has the highest permeability value of 1.574, but 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester has the least permeability valve of 0.927. Likewise, all the phytochemicals can be readily absorbed by the intestinal cells, with 2-Ethylacridine having the highest intestinal absorption of 98.202%. All the phytochemicals are substrate of P-glycoprotein expect 9,12-Octadecadienoic acid (Z,Z)-, cyclopentaneundecanoic acid, and o-Xylene. None of the phytochemicals is neither inhibitor of P-glycoprotein-I nor P-glycoprotein-II.
Table 18.
Predicted absorption properties of phytochemicals with best docking scores
| Ligand | Water solubility (log mol/L) | Caco2 permeability (log Papp in 10–6 cm/s) | Intestinal absorption (% absorbed) | Skin permeability (log Kp) | P-glycoprotein substrate | P-glycoprotein I inhibitor | P-glycoprotein II inhibitor |
|---|---|---|---|---|---|---|---|
| 9,12-Octadecadienoic acid (Z,Z)- | − 5.862 | 1.57 | 92.329 | − 2.723 | No | No | No |
| N-Cyclododecylacetamide | − 3.487 | 1.477 | 92.574 | − 2.178 | Yes | No | No |
| Cyclopentaneundecanoic acid | − 5.364 | 1.566 | 92.079 | − 2.716 | No | No | No |
| o-Xylene | − 2.552 | 1.574 | 96.713 | − 1.236 | No | No | No |
| 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester | − 3.763 | 0.964 | 88.393 | − 3.659 | Yes | No | No |
| 2-Ethylacridine | -5.037 | 1.355 | 98.202 | -2.246 | YES | NO | NO |
|
6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester |
-4.159 | 0.927 | 89.551 | -3.136 | YES | NO | NO |
| Cyclododecylamine | − 2.651 | 1.341 | 92.472 | − 2.402 | Yes | No | No |
| 1H-Indole, 5-methyl-2-phenyl- | − 5.006 | 1.541 | 93.388 | − 2.507 | Yes | No | No |
Predicted in vivo distribution, clearance, and cytochrome P450 promiscuity of the phytochemicals with best docking score
Table 19 shows the predicted in vivo distribution of the phytochemicals. All the phytochemicals tested have relatively low steady-state volume of distribution. Similarly, the predicted result demonstrated that Cyclododecylamine has the highest unbound fraction in the human blood. All the phytochemicals have relatively low blood–brain barrier and CNS permeability values.
Table 19.
Predicted in vivo distribution and excretion properties of phytochemicals with best docking scores
| Ligand | VDss (human) | Fraction unbound (human) | BBB permeability | CNS permeability | Total Clearance | Renal OCT2 substrate |
|---|---|---|---|---|---|---|
| 9,12-Octadecadienoic acid (Z,Z)- | − 0.587 | 0.054 | − 0.142 | − 1.6 | 1.936 | No |
| N-Cyclododecylacetamide | 0.277 | 0.49 | 0.494 | − 3.388 | 1.426 | No |
| Cyclopentaneundecanoic acid | − 0.568 | 0.084 | − 0.034 | − 1.567 | 1.465 | No |
| o-Xylene | 0.325 | 0.362 | 0.409 | − 1.677 | 0.269 | No |
| 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester | 0.119 | 0.324 | − 0.113 | − 3.032 | 0.996 | No |
| 2-Ethylacridine | 0.449 | 0.197 | 0.526 | − 1.372 | 0.829 | No |
|
6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester |
− 0.279 | 0.14 | − 0.332 | − 2.922 | 0.141 | No |
| Cyclododecylamine | 0.682 | 0.599 | 0.397 | − 3.167 | 0.707 | No |
| 1H-Indole, 5-methyl-2-phenyl- | 0.284 | 0.106 | 0.571 | − 1.384 | 0.417 | No |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | − 5463 | 1.791 | 98.046 | − 2.41 | 0.137 | No |
|
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
− 2.895 | 1.284 | 92.921 | − 3.189 | 0.481 | No |
The predicted clearance of each of the phytochemicals showed that 9,12-Octadecadienoic acid (Z,Z)- has the highest total clearance rate of 1.936, while 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane has the least clearance rate of − 0.137. None of the phytochemicals is substrate of renal organic cation transporter.
Table 20 displays the predicted human cytochrome P450 promiscuity of the phytochemicals. All the phytocompounds were neither substrate nor inhibitor of CYP2D6 expect 2-Ethylacridine which could be inhibitor of CYP2D6. Similarly, none of the phytochemicals expect cyclopentaneundecanoic acid is inhibitor of CYP3A4. Cyclododecylamine and 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester were neither substrate nor inhibitor of any of the cytochrome P450 tested in this study.
Table 20.
Predicted cytochrome P450 promiscuity of phytochemicals with best docking scores
| Ligand | CYP2D6 substrate | CYP3A4 substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor |
|---|---|---|---|---|---|---|---|
| 9,12-Octadecadienoic acid (Z,Z)- | No | Yes | Yes | No | No | No | No |
| N-Cyclododecylacetamide | No | No | No | No | No | No | No |
| Cyclopentaneundecanoic acid | No | No | Yes | No | No | No | Yes |
| o-Xylene | No | No | No | No | No | No | No |
| 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester | No | No | No | No | No | No | No |
| 2-Ethylacridine | No | YesS | Yes | Yes | Yes | Yes | No |
|
6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester |
No | YesS | No | No | No | No | No |
| Cyclododecylamine | No | No | No | No | No | No | No |
| 1H-Indole, 5-methyl-2-phenyl- | No | Yes | Yes | Yes | Yes | No | No |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | No | Yes | Yes | Yes | No | No | No |
|
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
No | No | Yes | No | No | No | No |
Predicted toxicological profile of the phytochemicals with best docking score
Table 21 displays the predicted toxicological profile of the phytochemicals with best docking scores with the pathogenic proteins. All the phytochemicals have no mutagenic potentials against bacteria (AMES toxicity) except 2-Ethylacridine and 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane could be toxic to bacteria. None of the phytochemicals has adverse effects on the hepatic cells expect 9,12-Octadecadienoic acid (Z,Z)-; 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester; and 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester. The phytochemicals are not inhibitors of human ether-a-go-go-related gene (hERG) hERG I and hERG II except 2-Ethylacridine and 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester which may inhibit hERG II. All the phytochemicals have relatively low maximum recommended tolerated dose values.
Table 21.
In silico toxicological properties of phytochemicals with best docking scores
| Ligand | Max. tolerated dose (human) | Oral rat acute toxicity (LD50) | Oral rat chronic toxicity (LOAEL) | T.Pyriformis toxicity | Minnow toxicity | AMES toxicity | hERG I inhibitor | hERG II inhibitor | Hepatotoxicity | Skin Sensitisation |
|---|---|---|---|---|---|---|---|---|---|---|
| 9,12-Octadecadienoic acid (Z,Z)- | − 0.827 | 1.429 | 3.187 | 0.701 | − 1.31 | No | No | No | Yes | Yes |
| N-Cyclododecylacetamide | 0.546 | 2.236 | 0.956 | 0.892 | 1.754 | No | No | No | No | Yes |
| Cyclopentaneundecanoic acid | − 0.762 | 1.559 | 3.027 | 0.812 | − 0.855 | No | No | No | No | Yes |
| o-Xylene | 0.921 | 1.841 | 2.169 | − 0.022 | 1.31 | No | No | No | No | No |
| 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester | − 0.35 | 2.548 | 0.34 | 0.417 | 1.616 | No | No | No | Yes | No |
| 2-Ethylacridine | − 0.255 | 1.739 | 1.146 | 1.188 | − 0.7 | Yes | No | Yes | No | No |
|
6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester |
2.712 | 0.99 | 1.052 | 1,393 | No | No | Yes | Yes | No | |
| Cyclododecylamine | 0.611 | 2,258 | 1.166 | 0.51 | 1.838 | No | No | No | No | Yes |
| 1H-Indole, 5-methyl-2-phenyl- | 0.147 | 1.904 | 1.002 | 1.274 | 0.554 | No | No | No | No | No |
| 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane | 0.149 | 2.468 | 2.065 | 1.151 | − 0.314 | Yes | No | No | No | No |
|
Carbamic acid, [1-(hydroxymethyl)-2-phenylethyl]-, 1,1-dimethylethyl ester, (s)- |
0.732 | 2.024 | 1.767 | 0.904 | 1.183 | No | No | No | No | No |
Discussion
SARS-CoV-2 has affected more than 212 million people with over 4.43 million fatalities globally, and the genome of this virus is associated with diverse mutation; hence, recognizing specific inherent proteins of this virus by small molecule drugs is a prime target in combatting its virulence (Kangarshahi et al. 2021; Patel et al. 2021). These proteins are of both structural (four) and non-structural (sixteen) functionality comprising approximately 7096 amino acid residues (Naqvi et al. 2020; Kumar et al. 2021). Hence, making multi-viral protein-targeting molecules suitable as potential anti-coronavirus drug candidates (Wu et al. 2020).
Oxidative stress, redox imbalance, cell death, inflammation, cytokine production, and other pathophysiological processes have been implicated in the pathogenesis of respiratory viral infections including severe acute respiratory syndrome coronavirus (SARSCoV) (Oladele et al. 2020a, b, c, d). Weakening antioxidant protective mechanisms and excessive secretion of reactive oxygen species (ROS) are vital for viral replication and the consequent virus-linked pathological conditions (Khomich et al. 2018). In this study, we reported the cytoprotective and antioxidative properties of six tropical plants used traditionally to prevent and treat COVID-19 and its associated clinical symptoms.
Several antioxidant assays were employed in this study to establish the antioxidant potentials of the aqueous extracts of the selected tropical plants. As shown in the results, all the extracts demonstrated substantial free radical scavenging and antioxidants activities. This indicates that these extracts serve as reservoirs of vital and medicinal phytochemicals such as phenols, flavonoids, tannins, and so on whose therapeutic efficacies have been scientifically established. These findings were corroborated by the GCMS analysis of the extracts revealing the phytochemicals present in them.
Scientific evidences have reported that activation of the oxidative stress machinery is vital in the initiation of severe lung injury in SARS-CoV-infected patients which associated with expression of transcription factors, including NF-kB, leading to an aggravated proinflammatory host response (Smith et al. 2012; Lin et al. 2006). Abrogation of this initiation process by phytochemicals present in extracts could provide health benefits and potent therapeutic solution to COVID-19 pandemic. Polyphenols and flavonoids have been reported in an in silico study to mitigate pathogenesis of COVID-19 (Oladele et al. 2021a, b).
It is well established that there exists link between oxidative stress and inflammation (Sies 2015). Activation of NF-kB with consequent excessive secretion proinflammatory cytokines is considered a crucial mediator in SARS-CoV pathophysiology (Smith et al. 2012; Padhan et al. 2008). In this study, the in vitro anti-inflammatory property of the extracts was assessed by evaluating their nitric oxide inhibitory effect. As depicted in the results, all the extracts exhibited significant nitric oxide inhibitory effect confirming their anti-inflammatory potentials. Nitric oxide is a mediator of nitrosative stress and also an inflammatory biomarker that has been implicated in pathogenesis of many diseases. Similarly, nitric oxide is mainly synthesized via inducible nitric oxide synthase (iNOS) under inflammatory conditions.
As shown in this present study, all the extracts contain phytochemicals that have inhibitory effect against the ADP ribose phosphatase. The macrodomain of the ADP ribose phosphatase has been documented to significantly contributes to the alteration of the viral-induced immune response of the host and enhance viral RNA replication owing to induced nuclear translocation of host type 1 interferon (INF-1) regulatory factor 3 (IRF3), an important innate immunity mediator. This property of seizing the host immune system makes this protein a unique drug target (Fehr et al. 2018; Michalska et al. 2020; Patel et al 2021). The in silico inhibition of this protein by the phytochemicals present in these extracts is indicative of their immune enhancing activity, thus playing vital role in mitigating the pathobiology of COVID-19.
Mainly associated with viral RNA replication and transcription, the RNA-dependent RNA polymerase (RdRp) in conjunction with other structural proteins such as Nsp 7 and Nsp 8 exhibits profound polymerase activity, making this enzyme a key target of most antiviral agents (Kirchdoerfer and Ward 2019; Jiang et al. 2021; Malone et al. 2022). In this study, phytochemicals present in these extracts remarkably inhibited RNA-dependent RNA polymerase as showed in the computational study, supporting the antiviral activity of these phytochemicals.
The result of this study also demonstrated that the phytochemicals contained in the aqueous extracts of the tropical plants markedly inhibited the SARS-CoV-2 main protease (Mpro). Mpro is a critical highly conserved protein engaged in the replication and maturation processes of SARS-CoV-2. It comprises of chymotrypsin and picornavirus 3 C protease-like catalytic domains. This protein is emerging as a significant antiviral target because of its role in proteolytic processing of viral polyproteins (Joshi et al. 2021; Abdul Amin et al. 2021).
Nsp9 and primase were also inhibited by the phytochemicals using computational approach. Nsp9, a 113 amino acid dimeric protein primarily functions in binding single stranded nucleic acids (DNA and RNA) during viral replication and its impairment, yields the synthesis of defective RNAs in the virus (Kangarshahi et al. 2021). Unlike other viral proteins/enzymes, the primase is vastly associated with de novo initiation of nucleic acids making it one of key enzymes driving the infectivity of SARS-CoV-2 (Konkolova et al. 2020). All these pathogenic proteins of COVID-19 were inhibited by the phytochemicals in the plant extracts suggesting them as potential anti-COVID-19 drug candidates.
The results of the solubility, pharmacodynamics, pharmacokinetics, and toxicological profiles of the phytochemicals were investigated as a systemic virtual screening of drugs and potential drugs. This is done as alternative to in vivo examinations which are essential complements in drug discovery. The Lipinski’s rule is a major criterion to evaluate drug likeliness and to determine if a compound with a particular pharmacological and biological action has physical and chemical properties that could favour its activities in human. The molecular properties of the compounds based on the computed partition coefficient (log P) showed that the phytochemicals have relatively good lipophilicity as the logP values were less than 5 (Lipinski et al. 1997; Hughes et al. 2008). All the tested phytochemicals could be maintained in the system at appropriate concentrations.
All the phytochemicals except 9,12-Octadecadienoic acid (Z,Z)-, cyclopentaneundecanoic acid, and o-Xylene were predicted to be substrates of P-glycoprotein, a member of the ATP-binding cassette transporter and an efflux membrane transporter found chiefly in epithelial cells. Conversely, none of the phytochemicals is predicted as P-glycoprotein inhibitors. This suggested that they don’t disrupt the normal physiological activities of P-glycoprotein including restricting the active uptake and the distribution of drugs (Srivalli and Lakshmi 2012).
Cytochrome P450 is a group of enzymes that perform crucial functions in drug metabolism. They play a major role in the activation of drugs and also in the toxicity effects of the drugs. None of the phytochemicals expect cyclopentaneundecanoic acid is inhibitor of CYP3A4. Also, all the phytocompounds were neither substrate nor inhibitor of CYP2D6 expect 2-Ethylacridine which could be inhibitor of CYP2D6. The lipophilicity of the drug appears to correlate negatively to metabolism-related toxicity. Furthermore, none of the phytochemicals were a substrate of renal organic cation transporter; this implies that they are possibly cleared through other available routes including bile, sweat, and so on. Furthermore, the phytochemical displayed substantial clearance results. Drug clearance is related to bioavailability and is crucial for determining dosing rates to achieve steady-state concentrations.
Acute and chronic toxicity were also carried out on the phytochemicals to determine the safety of the compounds when administered. Exposure to low-moderate doses/concentrations of xenobiotics over long period of time is of significant concern in many treatment strategies or interventions. Chronic studies are designed to identify the lowest dose of a compound that can result in adverse effects (LOAEL), and the highest dose at which no adverse effects are observed (NOAEL). The toxicological assessment of the phytochemicals revealed that all the phytochemicals are not hepatoxic expect 9,12-Octadecadienoic acid (Z,Z)-, 4-(2-Amino-3-cyano-5-oxo-5,6,7,8-tetrahydro-4H-chromen-4-yl)-3,5-dimethyl-1H-pyrrole-2-carboxylic acid ethyl esteryl) 3,5, dimethyl, 1H pyrrole-2-carboxylic acid ethyl ester, and 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrimidine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester. Similarly, only 1,3,3-Trimethyl-1-(4′-methoxyphenyl)-6-methoxyindane and 2-Ethylacridine are bacterial mutagenic potential drugs using the AMES toxicity examination. However, all the compounds showed high level of toxicity to Tetrahymena pyriformis toxicity test. None of the phytochemicals is an inhibitor to hERG I, but 2-Ethylacridine and 6-Methyl-2-oxo-4-(4-trifluoromethyl-phenyl)-1,2,3,4-tetrahydro-pyrim idine-5-carboxylic acid 2-methylsulfanyl-ethyl ester 2-methylsulfanyl ethyl ester may be inhibitors to hERG II. Inhibition of the hERG potassium channel could result in delayed ventricular repolarisation leading to a severe disturbance in the normal cardiac rhythm and disrupt hepatic functions (Oso et al. 2019; Oladele et al. 2021a, b).
Conclusion
Put together, the data from this study demonstrated that the phytochemicals of the selected tropical medicinal plants displayed substantial binding affinity to the binding pockets of four main pathogenic proteins of Coronavirus-2 indicating these phytochemicals as putative inhibitors of Coronavirus-2. The reaction between these phytocompounds and proteins of Coronavirus-2 could alter the pathophysiology of COVID-19, thus mitigating its pathogenic reactions/activities. This study reported the in silico inhibitory activity of phytochemicals against four pathogenic proteins of Coronavirus-2. However, in vitro and in vivo studies are important to potentiate these findings. Other drug techniques or models are vital to elucidate their compatibility and usage of the phytochemicals as adjuvants in vaccine development against the highly contagious COVID-19 infection.
Author contribution
Johnson Olaleye Oladele: Investigation, data curation; methodology; resources; writing—original draft. Taiwo Scholes Adewole: Investigation, data curation; methodology; resources; writing—original draft. Gbenga Emmanuel Ogundepo: Methodology, data curation, investigation. Oyedotun Moses Oyeleke: Data curation; investigation; methodology. Adenike Kuku: Conceptualization; supervision; project administration; resources; writing—review and editing.
Data availability
Data will be available upon request.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Amin SA, Banerjee S, Ghosh K, Gayen S, Jha T. Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg Med Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Sarmah S, Lyndem S, Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2021;39(9):3347–3357. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimzadeh MA, Nabavi SF, Nabavi SM. Antioxidant activities of methanol extract of Sambucus ebulus L. flower. Pak J Biol Sci: PJBS. 2009;12(5):447–450. doi: 10.3923/pjbs.2009.447.450. [DOI] [PubMed] [Google Scholar]
- Fehr AR, Jankevicius G, Ahel I, Perlman S. Viral macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol. 2018;26(7):598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan SS, Attrish D, Ghosh S, Choudhury PP, Roy B. Pathogenic perspective of missense mutations of ORF3a protein of SARS-CoV-2. Virus Res. 2021;300:198441. doi: 10.1016/j.virusres.2021.198441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Blagg J, Price DA, Bailey S, Decrescenzo GA, Devraj RV, Ellsworth E, Fobian YM, Gibbs ME, Gilles RW, Greene N, Huang E, Krieger-Burke T, Loesel J, Wager T, Whiteley L, Zhang Y (2008) [DOI] [PubMed]
- Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems. 2020;5(3):e00266–e320. doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yin W, Xu HE. RNA-dependent RNA polymerase: structure, mechanism, and drug discovery for COVID-19. Biochem Biophys Res Commun. 2021;538:47–53. doi: 10.1016/j.bbrc.2020.08.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, Pandya VK, Chugh A, Giri AP, Kulkarni MJ. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn. 2021;39(9):3099–3114. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangarshahi ZT, Lak S, Ghadam M, Motamed N, Sardari S, Rahimi S (2021) The proteins of SARS-CoV-2 and their functions.
- Khomich OA, Kochetkov SN, Bartosch B, et al. Redox biology of respiratory viral infections. Viruses. 2018;10:E392. doi: 10.3390/v10080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Minamikawa T. Hydroxyl radical-scavenging effects of spices and scavengers from brown mustard (Brassica nigra) Bios Biotechnol Biochem. 1997;61(1):118–123. doi: 10.1271/bbb.61.118. [DOI] [Google Scholar]
- Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkolova E, Klima M, Nencka R, Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J Struct Biol. 2020;211(2):107548. doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kashyap P, Chowdhury S, Kumar S, Panwar A, Kumar A. Identification of phytochemicals as potential therapeutic agents that binds to Nsp15 protein target of coronavirus (SARS-CoV-2) that are capable of inhibiting virus replication. Phytomedicine. 2021;85:153317. doi: 10.1016/j.phymed.2020.153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Lin KH, Hsieh TH, et al. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol Med Microbiol. 2006;46:375e380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW. Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Liu YW, Han CH, Lee MH, Hsu FL, Hou WC (2003) Patatin, the tuber storage protein of potato (Solanum tuberosum L.), exhibits antioxidant activity in vitro. J Agric Food Chem 51:4389–4393. 10.1021/jf030016j [DOI] [PubMed]
- Maio N, Lafont BA, Sil D, Li Y, Bollinger JM, Krebs C, Pierson TC, Linehan WM, Rouault TA. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat Rev Mol Cell Biol. 2022;23(1):21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K, Kim Y, Jedrzejczak R, Maltseva NI, Stols L, Endres M. Joachimiak A (2020) Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ, 7(5). [DOI] [PMC free article] [PubMed]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Computational Chemistry. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi SM, Ebrahimzadeh MA, Nabavi SF, Hamidinia A, Bekhradnia AR. Determination of antioxidant activity, phenol and flavonoids content of Parrotia persica Mey. Pharmacologyonline. 2008;2(9):560–567. [Google Scholar]
- Nabavi SM, Ebrahimzadeh MA, Nabavi SF, Fazelian M, Eslami B. In vitro antioxidant and free radical scavenging activity of Diospyros lotus and Pyrus boissieriana growing in Iran. Pharm Mag. 2009;5(18):122. [Google Scholar]
- Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, Atif SM, Hariprasad G, Hasan GM, Hassan MI. (2020) Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta (BBA)- Mol Basis Dis. 1866;10:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladele JO, Ajayi EIO, Oyeleke OM, Oladele OT, Olowookere BD, Adeniyi BM, Oyewole OI, Oladiji AT. A systematic review on COVID-19 pandemic with special emphasis on curative potentials of medicinal plants. Heliyon. 2020;6:1–17. doi: 10.1016/j.heliyon.2020.e04897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladele JO, Ajayi EIO, Oyeleke OM, Oladele OT, Olowookere BD, Adeniyi BM, Oyewole OI. Curative potentials of Nigerian medicinal plants in COVID-19 treatment: a mechanistic approach. Jordan Jo Biol Sci. 2020;13:681–700. [Google Scholar]
- Oladele JO, Oyeleke OM, Oladele OT, Olaniyan MD. Neuroprotective mechanism of Vernonia amygdalina in a rat model of neurodegenerative diseases. Toxicol rep. 2020;7:1223–1232. doi: 10.1016/j.toxrep.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladele JO, Oyeleke OM, Oladele OT, Oladiji AT. COVID-19 treatment: investigation on the phytochemical constituents of Vernonia amygdalina as potential coronavirus-2 inhibitors. Comput Toxicol. 2021;18:100161. doi: 10.1016/j.comtox.2021.100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladele JO, Anyim JC, Oyeleke OM, Olowookere BD, Bamigboye MO, Oladele OO, Oladiji AT. Telfairia occidentalis mitigates dextran sodium sulphate-induced ulcerative colitis in rats via suppression of oxidative stress, lipid peroxidation and inflammation. J Food Biochem. 2021 doi: 10.1111/jfbc.13873. [DOI] [PubMed] [Google Scholar]
- Oladele JO, Oyeleke OM, Awosanya OO, Olowookere BD, Oladele OT (2020c) Fluted Pumpkin (Telfaira occidentalis) protects against phenyl hydrazine-induced anaemia and toxicities in rats. Adv Tradit Med.
- Oso BJ, Oyewo EB, Oladiji AT. Influence of ethanolic extracts of dried fruit of Xylopia aethiopica (Dunal) A. Rich on haematological and biochemical parameters in healthy Wistar rats. Clin Phytoscience. 2019;5(1):9. doi: 10.1186/s40816-019-0104-4. [DOI] [Google Scholar]
- Padhan K, Minakshi R, Towheed MAB, et al. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol. 2008;89:1960e1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Patel DC, Hausman KR, Arba M, Tran A, Lakernick PM, Wu C (2021) Novel inhibitors to ADP ribose phosphatase of SARS-CoV-2 identified by structure-based high throughput virtual screening and molecular dynamics simulations. Comput biol med, p.105084. [DOI] [PMC free article] [PubMed]
- Shahi Z, Sayyed-Alangi SZ, Najafian L. Effects of enzyme type and process time on hydrolysis degree, electrophoresis bands and antioxidant properties of hydrolyzed proteins derived from defatted Bunium persicum Bioss. press cake. Heliyon. 2020;6(2):e03365. doi: 10.1016/j.heliyon.2020.e03365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015;4:180e183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT, Willey NJ, Hancock JT. Low dose ionizing radiation produces too few reactive oxygen species to directly affect antioxidant concentrations in cells. Biol Lett. 2012;8:594e597. doi: 10.1098/rsbl.2012.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivalli KM, Lakshmi PK. Overview of P-glycoprotein inhibitors: a rational outlook. Braz J Pharm Sci. 2012;48(3):353–367. doi: 10.1590/S1984-82502012000300002. [DOI] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shen Y, Li M, Chuang H, Ye Y, Zhao H, Wang H (2020) Clinical manifestations and evidence of neurological involvement in 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J. Neurol. 1–13. [DOI] [PMC free article] [PubMed]
- Wu HC, Chen HM, Shiau CY. Free amino acids and peptides as related to antioxidant properties in protein hydrolysates of mackerel (Scomber austriasicus) Food Res Int. 2003;36(9–10):949–957. doi: 10.1016/S0963-9969(03)00104-2. [DOI] [Google Scholar]
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request.