Abstract
The emergence and rapid spread of carbapenemase‐producing Enterobacterales represents a serious public health concern. Critically, these global priority bacteria have begun to be reported in companion animals, implying a potential risk of cross‐transmission between humans and pets. Using long‐read (MinION) and short‐read (Illumina) sequencing technologies, we have identified and characterized a hypermucoviscous KPC‐2‐producing Klebsiella pneumoniae strain belonging to the high‐risk international clone ST11/CG258, in a dog with urinary tract infection. Strikingly, the bla KPC‐2 gene was carried by a 54‐kb IncN plasmid assignated to ST15, which shared 99.8 and 96.8% pairwise identity with IncN‐pST15 plasmids from human and environmental K. pneumoniae strains, respectively; all come from an area with high endemicity of KPC‐2. Our findings suggest that IncN‐pST15 plasmids conferring carbapenem resistance can play as important a role as clonal transmission of K. pneumoniae, representing another major challenge for One Health.
Keywords: carbapenemase, global priority pathogens, one health, pets, plasmidome
1. INTRODUCTION
Epidemiological studies have revealed that carbapenemase‐producing Enterobacterales have emerged in healthy and sick animals, and community settings (Kelly et al., 2017; Wang et al., 2020; Zhang et al., 2019), implying a potential risk of transmission of these pathogens between humans and companion animals (Grönthal et al., 2018; Sellera & Lincopan, 2019). Additionally, the transfer of carbapenems resistance genes can be facilitated by mobile genetic elements (e.g. plasmids and transposons), which is a concerning possibility (Baquero et al., 2019; Brandt et al., 2019).
KPC family has been the most widespread of all carbapenemases associated with Enterobacterales (van Duin & Doi, 2017). The occurrence of KPC‐producing bacteria in human hospital settings has rendered nosocomial infections particularly difficult to treat or even untreatable (Wang et al., 2016). To date, the identification of KPC producers in companion animals has been sporadically reported from dogs in Brazil (KPC‐2‐producing Escherichia coli) and United States (KPC‐4‐producing Enterobacter xiangfangensis) (Daniels et al., 2018; Sellera et al., 2018).
In this study, under a ‘One Health’ view, we report the identification of a KPC‐2‐positive Klebsiella pneumoniae belonging to the international high‐risk clone sequence type 11/clonal group 258 (ST11/CG258) in a dog suffering from urinary tract infection, highlighting that IncN‐pST15 plasmids carrying bla KPC‐2 genes are spreading among human, animal and environmental clonally unrelated K. pneumoniae strains.
2. MATERIALS AND METHODS
In 2019, during a Brazilian surveillance study (OneBR project), conducted to characterize the burden of antimicrobial resistance associated with critical WHO priority pathogens, a carbapenem‐resistant K. pneumoniae strain (PVT01) identified by BD Phoenix (BD Diagnostics, Sparks, MD, USA) was isolated from a urine culture of a 9‐year‐old female Spitz dog suffering from urinary tract infection.
Antimicrobial susceptibility testing was performed by the disc diffusion and/or Etest methods according to Clinical and Laboratory Standards Institute methods (CLSI, 2018, 2020). The antibiotics tested were amoxicillin/clavulanic acid, aztreonam, cefotaxime, ceftriaxone, cefepime, cefoxitin, ceftiofur, ciprofloxacin, enrofloxacin, nalidixic acid, chloramphenicol, amikacin, gentamicin, ertapenem, imipenem, meropenem, sulfamethoxazole/trimethoprim and tetracycline. Colistin susceptibility testing was performed by broth microdilution method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2021) guidelines. ESBL production was screened by the double‐disc synergy test (DDST) (Drieux et al., 2008), whereas phenotypic detection of KPC enzyme was performed by the combined disc test using imipenem disc supplemented with aminophenylboronic acid (Tsakris et al., 2011). In addition, PVT01 strain was screened for hypermucoviscosity by string test (Shon et al., 2013).
Total genomic DNA was extracted and sequenced using long‐read (MinION, Oxford Nanopore) and short‐read (NextSeq, Illumina) sequencing technologies. Hybrid de novo assembly was performed using Unicycler v0.4.8 (https://github.com/rrwick/Unicycler), whereas Mlplasmids (https://sarredondo.shinyapps.io/mlplasmids/) was used to predict plasmid and chromosome‐derived sequences (Arredondo‐Alonso et al., 2018). Genome sequences were annotated with NCBI PGAP v.3.2 (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/). ABRicate v0.9.8 (https://github.com/tseemann/abricate), with ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) and PlasmidFinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) databases, and Kleborate (https://github.com/katholt/Kleborate) were used for prediction of resistome, plasmidome, species confirmation, multilocus sequence type (ST), virulence loci, and K (capsule) and O antigen (LPS) serotypes (Lam et al., 2018; Wick et al., 2018; Wyres et al., 2016). The nucleotide sequences of K. pneumoniae strain PVT01 were deposited at GenBank under accession number JABSUB000000000.1.
3. RESULTS AND DISCUSSION
The PVT01 strain exhibited a multidrug‐resistant (MDR) profile (Magiorakos et al., 2012) to amoxicillin/clavulanic acid, aztreonam, ceftriaxone, ceftazidime, cefoxitin, cefotaxime, cefepime, ceftiofur, ertapenem, imipenem, meropenem, amikacin, gentamicin, sulfamethoxazole/trimethoprim, enrofloxacin, ciprofloxacin, nalidixic acid and chloramphenicol, remaining susceptible to tetracycline and colistin (Table 1). ESBL and carbapenemase production were confirmed by the phenotypic tests. Additionally, the PVT01 strain displayed a hypermucoviscous phenotype, as defined by a positive string test (i.e. viscous filament ≥ 5 mm in length).
TABLE 1.
Susceptibility profile and genomic features of KPC‐2‐producing Klebsiella pneumoniae strain isolated from an infected dog in Brazil
| Susceptibility profile a | |
| Amoxicillin/clavulanic acid | R |
| Aztreonam | R |
| Cefotaxime | R |
| Ceftriaxone | R |
| Ceftazidime | R |
| Ceftiofur | R |
| Cefoxitin | R |
| Cefepime | R |
| Ertapenem | R |
| Imipenem (MIC mg/L) | R (>32) |
| Meropenem (MIC mg/L) | R (>32) |
| Amikacin (MIC mg/L) | R (64) |
| Gentamicin (MIC mg/L) | R (>256) |
| Sulfamethoxazole/trimethoprim | R |
| Nalidixic acid | R |
| Enrofloxacin | R |
| Ciprofloxacin | R |
| Chloramphenicol | R |
| Tetracycline | S |
| Colistin (MIC mg/L) | S (2) |
| Molecular epidemiology | |
| MLST (ST/CG) b | 11/258 |
| K‐locus | KL15 |
| wzi | 50 |
| Serotype | O4 |
| Resistome | |
| β‐lactams | bla KPC−2, bla CTX‐M−15, bla LAP−2, bla OXA−1, bla SHV−11 |
| Quinolones | aac(6')‐Ib‐cr, oqxA, oqxB, qnrS1, gyrA (S83I), parC (S80I) |
| Aminoglycosides | aac(3)‐IIa, aadA2, aph(3')‐Ia |
| Sulfamethoxazole | sul1 |
| Trimethoprim | dfrA12 |
| Fosfomycin | fosA |
| Macrolides | mphA |
| Chloramphenicol | catB4 |
| Virulome | |
| Yersiniabactin siderophore | ybt, fyuA, irp |
| Plasmidome | |
| Inc‐type [size, kb] c | IncFIB(K) [168], IncN [54], Col4401‐like [76] |
| GenBank accession number | JABSUB000000000.1 |
Susceptibility profiles were determined using the CLSI guideline (CLSI, 2020). For ceftiofur, enrofloxacin and colistin, resistance profiles were determined using veterinary CLSI (CLSI, 2018) and EUCAST 2021 (https://www.eucast.org/) guidelines, respectively.
MLST, Multi‐Locus Sequence Typing; ST, sequence type; CG, clonal group.
The IncFIB(K) plasmid, named pPVT01_P1, harboured bla CTX‐M‐15, aac(3)‐IIa, aadA2, aph(3')‐Ia, mphA, sul and dfrA12, whereas Col4401‐like plasmid (pPVT01_P2) harboured bla OXA‐1, bla LAP‐2, qnrS1 and aac(6')‐Ib‐cr resistance genes.
Resistome analysis revealed a MDR genotype to β‐lactams, quinolones, aminoglycosides, sulfamethoxazole/trimethoprim, fosfomycin, macrolides and chloramphenicol (Table 1). Moreover, genes encoding for yersiniabactin siderophore synthesis (ybt, fyuA and irp genes) (Paczosa & Mecsas, 2016), and KL15 (wzi50) and O4 loci were identified (Wyres et al., 2016).
Hybrid assembly revealed three resistance plasmids: IncFIB(K) (168‐kb), IncN (54‐kb) and Col4401‐like (76‐kb). The IncFIB(K) plasmid, named pPVT01_P1, harboured bla CTX‐M‐15, aac(3)‐IIa, aadA2, aph(3')‐Ia, mphA, sul1 and dfrA12, whereas Col4401(76‐kb)‐like plasmid (pPVT01_P2) harboured bla OXA‐1, bla LAP‐2, qnrS1 and aac(6')‐Ib‐cr resistance genes. Specifically, the bla KPC‐2 gene was carried by the 54‐kb IncN plasmid (named pPVT01_P3) assignated to ST15 by pMLST typing and located on a Tn4401 transposon > 99% identical to Tn4401b isoform (GenBank accession number: EU176012). The plasmid pPVT01_P3 (GenBank accession number: JABSUB010000003.1) shared 99.8 and 96.8% pairwise identity with pKPC_FCF/3SP and pKP148 IncN‐pST15plasmids (GenBank accession numbers: CP004367.2 and KX062091.1), previously identified in human and environmental K. pneumoniae strains belonging to ST442 (Pérez‐Chaparro et al., 2014) and ST437, respectively (Oliveira et al., 2014) (Figure 1). Strikingly, all these K. pneumoniae strains come from an area with high endemicity of KPC‐2 (Sampaio & Gales, 2016), highlighting the widespread and adaptation of IncN‐pST15 plasmids carrying bla KPC‐2 at the human–animal–environment interface (Rada et al., 2020), and addressing a One‐Health implication to the problem of rapid dissemination of KPC‐2‐producing K. pneumoniae. In fact, K. pneumoniae PVT01 belonged to ST11/CG258, recognized as an international high‐risk clone linked to the epidemiological success of pandemic KPC carbapenemases in nosocomial settings (Bialek‐Davenet et al., 2014; Kelly et al., 2017; Rojas et al., 2017; Wyres & Holt, 2018). Worryingly, adaptation of ST11 to veterinary settings has been documented in European and Asian countries (Donati et al., 2014; Hidalgo et al., 2013; Loncaric et al., 2016; Mairi et al., 2020; Ovejero et al., 2017; Pilo et al., 2015; Schmidt et al., 2020; Wang et al., 2020; Wohlwend et al., 2015; Zhang et al., 2019), with KPC‐2‐positive ST11 only being reported in horse (Wang et al., 2020) and swine (Zhang et al., 2019) in China, so far.
FIGURE 1.
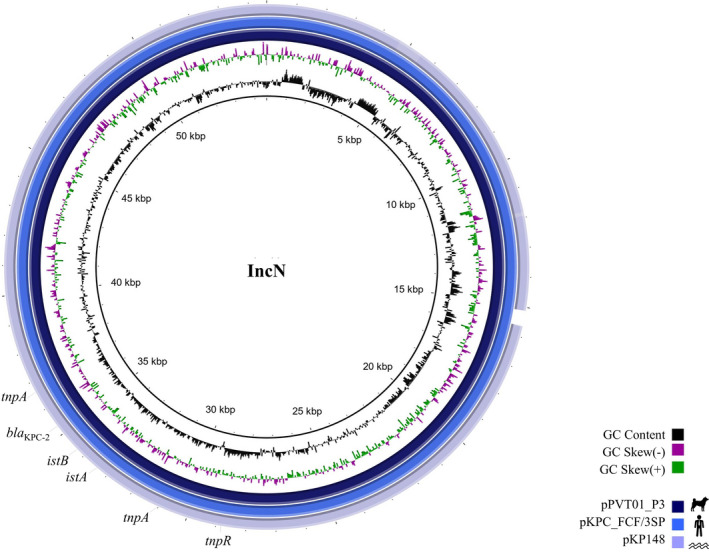
BRIG comparative analysis of pPVT01_P3 IncN‐pST15 plasmid harbouring bla KPC‐2, from a Klebsiella pneumoniae belonging to ST11/CG258 isolated from a dog suffering from urinary tract infection, with two closely related bla KPC‐2‐positive IncN‐pST15 plasmids from human (pKPC_FCF/3SP, GenBank accession number: CP004367.2) and environmental (pKP148 plasmid, GenBank accession number: KX062091.1) K. pneumoniae strains. The coloured rings denote similarity between the plasmid sequences
In summary, to the best of our knowledge, this is the first report of KPC‐positive K. pneumoniae ST11/CG258 isolated from a pet. The emergence of KPC‐2‐producing bacteria in companion animals is an important public health issue that denotes that pets are a neglected reservoir for critical priority pathogens in the community, and susceptible hosts for acquisition of untreatable or difficult‐to‐treat infections (Abraham et al., 2014; Köck et al., 2018; Pomba et al., 2017; Sellera & Lincopan, 2019). In this regard, IncN‐pST15 plasmids conferring carbapenem resistance can play as important a role as clonal transmission of K. pneumoniae, representing another major challenge for One Health. Therefore, surveillance studies should investigate similarities of plasmids circulating at the human–environment–animal interface in addition to clonal transmission.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
ETHICAL APPROVAL
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required for this specific study.
ACKNOWLEDGEMENTS
This work was funded by research grants from Bill & Melinda Gates Foundation, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). N. L. is a research fellow of CNPq (312249/2017‐9). We thank Cefar Diagnóstica Ltda. (São Paulo, Brazil) and CEFAP‐GENIAL facility for kindly supplying antibiotic discs for susceptibility testing and Illumina sequencing, respectively.
Sellera FP, Fuga B, Fontana H, et al. Detection of IncN‐pST15 one‐health plasmid harbouring bla KPC‐2 in a hypermucoviscous Klebsiella pneumoniae CG258 isolated from an infected dog, Brazil. Transbound Emerg Dis. 2021;68:3083–3088. 10.1111/tbed.14006
Fábio P. Sellera and Bruna Fuga equally contributed to this article.
FUNDING
Bill & Melinda Gates Foundation [Grand Challenges Explorations Brazil—New approaches to characterize the global burden of antimicrobial resistance, Grant OPP1193112]; Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP, Grant 2016/08593‐9]; Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq Grants 443819/2018‐1, 312249/2017‐9 and 433128/2018‐6]; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [CAPES Grant 88,887.358057/2019‐00].
Contributor Information
Fábio P. Sellera, Email: fsellera@usp.br.
Nilton Lincopan, Email: lincopan@usp.br.
DATA AVAILABILITY STATEMENT
All data generated or used during the study appear in the submitted article. The data that support the findings of this study are available from the corresponding author upon reasonable request. The whole genome nucleotide sequence of the K. pneumoniae PVT01 strain is available in the GenBank database under accession number JABSUB000000000.1. Genomic data of K. pneumoniae strain PVT01 is also available on the OneBR platform (http://onehealthbr.com/) under the number ID ONE247.
REFERENCES
- Abraham, S. , Wong, H. S. , Turnidge, J. , Johnson, J. R. , & Trott, D. J. (2014). Carbapenemase‐producing bacteria in companion animals: A public health concern on the horizon. Journal of Antimicrobial Chemotherapy, 69, 1155–1157. 10.1093/jac/dkt518 [DOI] [PubMed] [Google Scholar]
- Arredondo‐Alonso, S. , Rogers, M. R. C. , Braat, J. C. , Verschuuren, T. D. , Top, J. , Corander, J. , Willems, R. J. L. , & Schürch, A. C. (2018). Mlplasmids: A user‐friendly tool to predict plasmid‐and chromosome‐derived sequences for single species. Microbial Genomics, 4, e000224. 10.1099/mgen.0.000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero, F. , Coque, T. M. , Martínez, J. L. , Aracil‐Gisbert, S. , & Lanza, V. F. (2019). Gene transmission in the One Health microbiosphere and the channels of antimicrobial resistance. Frontiers in Microbiology, 10, 2892. 10.3389/fmicb.2019.02892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek‐Davenet, S. , Criscuolo, A. , Ailloud, F. , Passet, V. , Jones, L. , Delannoy‐Vieillard, A. S. , & Brisse, S. (2014). Genomic definition of hypervirulent and multidrug‐resistant Klebsiella pneumoniae clonal groups. Emerging Infectious Diseases, 20, 1812–1820. 10.3201/eid2011.140206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, C. , Viehweger, A. , Singh, A. , Pletz, M. W. , Wibberg, D. , Kalinowski, J. , Lerch, S. , Müller, B. , & Makarewicz, O. (2019). Assessing genetic diversity and similarity of 435 KPC‐carrying plasmids. Scientific Reports, 9, 11223. 10.1038/s41598-019-47758-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2018). Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. CLSI Supplement VET08, 4th ed. CLSI. [Google Scholar]
- Clinical and Laboratory Standards Institute (2020). Performance standards for antimicrobial susceptibility testing: Fifteenth informational supplement. CLSI document M100‐S30. CLSI. [Google Scholar]
- Daniels, J. B. , Chen, L. , Grooters, S. V. , Mollenkopf, D. F. , Mathys, D. A. , Pancholi, P. , & Wittum, T. E. (2018). Enterobacter cloacae complex sequence type 171 isolates expressing KPC‐4 carbapenemase recovered from canine patients in Ohio. Antimicrobial Agents and Chemotherapy, 62, e01161–e1218. 10.1128/AAC.01161-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati, V. , Feltrin, F. , Hendriksen, R. S. , Svendsen, C. A. , Cordaro, G. , García‐Fernández, A. , Lorenzetti, S. , Lorenzetti, R. , Battisti, A. , & Franco, A. (2014). Extended‐spectrum‐beta‐lactamases, AmpC beta‐lactamases and plasmid mediated quinolone resistance in Klebsiella spp. from companion animals in Italy. PLoS One, 9, e90564. 10.1371/journal.pone.0090564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drieux, L. , Brossier, F. , Sougakoff, W. , & Jarlier, V. (2008). Phenotypic detection of extended‐spectrum beta‐lactamase production in Enterobacteriaceae: Review and bench guide. Clinical Microbiology and Infection, 4(Suppl 1), 90–103. 10.1111/j.1469-0691.2007.01846.x [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (2021). Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. Retrieved from https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf
- Grönthal, T. , Österblad, M. , Eklund, M. , Jalava, J. , Nykäsenoja, S. , Pekkanen, K. , & Rantala, M. (2018). Sharing more than friendship ‐ transmission of NDM‐5 ST167 and CTX‐M‐9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Eurosurveillance, 23, 1700497. 10.2807/15607917.ES.2018.23.27.1700497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo, L. , Gutierrez, B. , Ovejero, C. M. , Carrilero, L. , Matrat, S. , Saba, C. K. S. , Santos‐Lopez, A. , Thomas‐Lopez, D. , Hoefer, A. , Suarez, M. , Santurde, G. , Martin‐Espada, C. , & Gonzalez‐Zorn, B. (2013). Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA‐1 β‐lactamase, and QnrB4. Antimicrobial Agents and Chemotherapy, 57, 4532–4534. 10.1128/AAC.00491-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A. M. , Mathema, B. , & Larson, E. L. (2017). Carbapenem‐resistant Enterobacteriaceae in the community: A scoping review. International Journal of Antimicrobial Agents, 50, 127–134. 10.1016/j.ijantimicag.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck, R. , Daniels‐Haardt, I. , Becker, K. , Mellmann, A. , Friedrich, A. W. , Mevius, D. , Schwarz, S. , & Jurke, A. (2018). Carbapenem‐resistant Enterobacteriaceae in wildlife, food‐producing, and companion animals: A systematic review. Clinical Microbiology and Infection, 24, 1241–1250. 10.1016/j.cmi.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Lam, M. M. C. , Wick, R. R. , Wyres, K. L. , Gorrie, C. L. , Judd, L. M. , Jenney, A. W. J. , Brisse, S. , & Holt, K. E. (2018). Genetic diversity, mobilisation and spread of the yersiniabactin‐encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microbial Genomics, 4, e000196. 10.1099/mgen.0.000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncaric, I. , Beiglböck, C. , Feßler, A. T. , Posautz, A. , Rosengarten, R. , Walzer, C. , Ehricht, R. , Monecke, S. , Schwarz, S. , Spergser, J. , & Kübber‐Heiss, A. (2016). Characterization of ESBL‐ and AmpC‐producing and fluoroquinolone‐resistant Enterobacteriaceae isolated from Mouflons (Ovis orientalis musimon) in Austria and Germany. PLoS One, 11, e0155786. 10.1371/journal.pone.0155786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos, A.‐P. , Srinivasan, A. , Carey, R. B. , Carmeli, Y. , Falagas, M. E. , Giske, C. G. , Harbarth, S. , Hindler, J. F. , Kahlmeter, G. , Olsson‐Liljequist, B. , Paterson, D. L. , Rice, L. B. , Stelling, J. , Struelens, M. J. , Vatopoulos, A. , Weber, J. T. , & Monnet, D. L. (2012). Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18, 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mairi, A. , Barraud, O. , Muggeo, A. , de Champs, C. , & Touati, A. (2020). Genomic analysis of a multidrug‐resistant Klebsiella pneumoniae ST11 strain recovered from Barbary deer (Cervus elaphus barbarus) in Akfadou Forest, Algeria. Journal of Global Antimicrobial Resistance, 22, 515–518. 10.1016/j.jgar.2020.04.027 [DOI] [PubMed] [Google Scholar]
- Oliveira, S. , Moura, R. A. , Silva, K. C. , Pavez, M. , McCulloch, J. A. , Dropa, M. , Matte, M. H. , Mamizuka, E. M. , Sato, M. I. Z. , Pestana de Castro, A. F. , & Lincopan, N. (2014). Isolation of KPC‐2‐producing Klebsiella pneumoniae strains belonging to the high‐risk multiresistant clonal complex 11 (ST437 and ST340) in urban rivers. Journal of Antimicrobial Chemotherapy, 69, 849–852. 10.1093/jac/dkt431 [DOI] [PubMed] [Google Scholar]
- Ovejero, C. M. , Escudero, J. A. , Thomas‐Lopez, D. , Hoefer, A. , Moyano, G. , Montero, N. , Martin‐Espada, C. , & Gonzalez‐Zorn, B. (2017). Highly tigecycline‐resistant Klebsiella pneumoniae sequence type 11 (ST11) and ST147 isolates from companion animals. Antimicrobial Agents and Chemotherapy, 61, e02640–e2716. 10.1128/AAC.02640-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczosa, M. K. , & Mecsas, J. (2016). Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiology and Molecular Biology Reviews, 80, 629–661. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Chaparro, P. J. , Cerdeira, L. T. , Queiroz, M. G. , de Lima, C. P. S. , Levy, C. E. , Pavez, M. , Lincopan, N. , Goncalves, E. C. , Mamizuka, E. M. , Sampaio, J. L. M. , Nunes, M. R. T. , & McCulloch, J. A. (2014). Complete nucleotide sequences of two bla KPC‐2‐bearing IncN plasmids isolated from sequence type 442 Klebsiella pneumoniae clinical strains four years apart. Antimicrobial Agents and Chemotherapy, 58, 2958–2960. 10.1128/AAC.02341-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo, P. , Vogt, D. , Origgi, F. C. , Endimiani, A. , Peterson, S. , & Perreten, V. (2015). First report of a multidrug‐resistant Klebsiella pneumoniae of sequence type 11 causing sepsis in a free‐ranging beaver (Castor fiber). Environmental Microbiology Reports, 7, 351–353. 10.1111/1758-2229.12257 [DOI] [PubMed] [Google Scholar]
- Pomba, C. , Rantala, M. , Greko, C. , Baptiste, K. E. , Catry, B. , van Duijkeren, E. , Mateus, A. , Moreno, M. A. , Pyörälä, S. , Ružauskas, M. , Sanders, P. , Teale, C. , Threlfall, E. J. , Kunsagi, Z. , Torren‐Edo, J. , Jukes, H. , & Törneke, K. (2017). Public health risk of antimicrobial resistance transfer from companion animals. Journal of Antimicrobial Chemotherapy, 72, 957–968. 10.1093/jac/dkw481 [DOI] [PubMed] [Google Scholar]
- Rada, A. M. , De La Cadena, E. , Agudelo, C. , Capataz, C. , Orozco, N. , Pallares, C. , & Restrepo, E. (2020). Dynamics of bla KPC‐2 dissemination from non‐CG258 Klebsiella pneumoniae to other Enterobacterales via IncN plasmids in an area of high endemicity. Antimicrobial Agents and Chemotherapy, 64, e01743–e1820. 10.1128/AAC.01743-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, L. J. , Weinstock, G. M. , De La Cadena, E. , Diaz, L. , Rios, R. , Hanson, B. M. , Brown, J. S. , Vats, P. , Phillips, D. S. , Nguyen, H. , Hujer, K. M. , Correa, A. , Adams, M. D. , Perez, F. , Sodergren, E. , Narechania, A. , Planet, P. J. , Villegas, M. V. , Bonomo, R. A. , & Arias, C. A. (2017). An analysis of the epidemic of Klebsiella pneumoniae carbapenemase‐producing K. pneumoniae: Convergence of two evolutionary mechanisms creates the "perfect storm". Journal of Infectious Diseases, 217, 82–92. 10.1093/infdis/jix524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio, J. L. , & Gales, A. C. (2016). Antimicrobial resistance in Enterobacteriaceae in Brazil: Focus on β‐lactams and polymyxins. The Brazilian Journal of Microbiology, 47, 31–37. 10.1016/j.bjm.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J. S. , Kuster, S. P. , Nigg, A. , Dazio, V. , Brilhante, M. , Rohrbach, H. , Bernasconi, O. J. , Büdel, T. , Campos‐Madueno, E. I. , Gobeli Brawand, S. , Schuller, S. , Endimiani, A. , Perreten, V. , & Willi, B. (2020). Poor infection prevention and control standards are associated with environmental contamination with carbapenemase‐producing Enterobacterales and other multidrug‐resistant bacteria in Swiss companion animal clinics. Antimicrobial Resistance and Infection Control, 9, 93. 10.1186/s13756-020-00742-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellera, F. P. , Fernandes, M. R. , Ruiz, R. , Falleiros, A. C. M. , Rodrigues, F. P. , Cerdeira, L. , & Lincopan, N. (2018). Identification of KPC‐2‐producing Escherichia coli in a companion animal: A new challenge for veterinary clinicians. Journal of Antimicrobial Chemotherapy, 73, 2259–2261. 10.1093/jac/dky173 [DOI] [PubMed] [Google Scholar]
- Sellera, F. P. , & Lincopan, N. (2019). Zooanthroponotic transmission of high‐risk multidrug‐resistant pathogens: A neglected public health issue. Journal of Infection and Public Health, 12, 294–295. 10.1016/j.jiph.2018.12.013 [DOI] [PubMed] [Google Scholar]
- Shon, A. S. , Bajwa, R. P. , & Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence, 4, 107–118. 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakris, A. , Themeli‐Digalaki, K. , Poulou, A. , Vrioni, G. , Voulgari, E. , Koumaki, V. , Agodi, A. , Pournaras, S. , & Sofianou, D. (2011). Comparative evaluation of combined‐disk tests using different boronic acid compounds for detection of Klebsiella pneumoniae carbapenemase‐producing Enterobacteriaceae clinical isolates. Journal of Clinical Microbiology, 49, 2804–2809. 10.1128/JCM.00666-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin, D. , & Doi, Y. (2017). The global epidemiology of carbapenemase‐producing Enterobacteriaceae . Virulence, 8, 460–469. 10.1080/21505594.2016.1222343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Li, X. , & Liu, B. T. (2020). Occurrence and characterization of KPC‐2‐producing ST11 Klebsiella pneumoniae isolate and NDM‐5‐producing Escherichia coli isolate from the same horse of equestrian clubs in China. Transboundary and Emerging Diseases, [Epub ahead of print]. 10.1111/tbed.13614 [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Zhang, Y. , Yao, X. , Xian, H. , Liu, Y. , Li, H. , Chen, H. , Wang, X. , Wang, R. , Zhao, C. , Cao, B. , & Wang, H. (2016). Risk factors and clinical outcomes for carbapenem‐resistant Enterobacteriaceae nosocomial infections. European Journal of Clinical Microbiology and Infectious Diseases, 35, 1679–1689. 10.1007/s10096-016-2710-0 [DOI] [PubMed] [Google Scholar]
- Wick, R. R. , Heinz, E. , Holt, K. E. , & Wyres, K. L. (2018). Kaptive web: User‐friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. Journal of Clinical Microbiology, 56, e00197–e218. 10.1128/JCM.00197-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlwend, N. , Endimiani, A. , Francey, T. , & Perreten, V. (2015). Third‐generation‐cephalosporin‐resistant Klebsiella pneumoniae isolates from humans and companion animals in Switzerland: Spread of a DHA‐producing sequence type 11 clone in a veterinary setting. Antimicrobial Agents and Chemotherapy, 59, 2949–2955. 10.1128/AAC.04408-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres, K. L. , & Holt, K. E. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Current Opinion in Microbiology, 45, 131–139. 10.1016/j.mib.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Wyres, K. L. , Wick, R. R. , Gorrie, C. , Jenney, A. , Follador, R. , Thomson, N. R. , & Holt, K. E. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microbial Genomics, 2, e000102. 10.1099/mgen.0.000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Zhu, Y. , Wang, C. , Liu, W. , Li, R. , Chen, F. , & Liu, S. (2019). Characterization of a multidrug‐resistant porcine klebsiella pneumoniae sequence type 11 strain coharboring blaKPC‐2 and fosA3 on two novel hybrid plasmids. mSphere, 4, e00590–e619. 10.1128/mSphere.00590-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or used during the study appear in the submitted article. The data that support the findings of this study are available from the corresponding author upon reasonable request. The whole genome nucleotide sequence of the K. pneumoniae PVT01 strain is available in the GenBank database under accession number JABSUB000000000.1. Genomic data of K. pneumoniae strain PVT01 is also available on the OneBR platform (http://onehealthbr.com/) under the number ID ONE247.


