Extended Data Fig. 10. Analysis of ubiquitous and abundant exosomal proteins using different approaches.
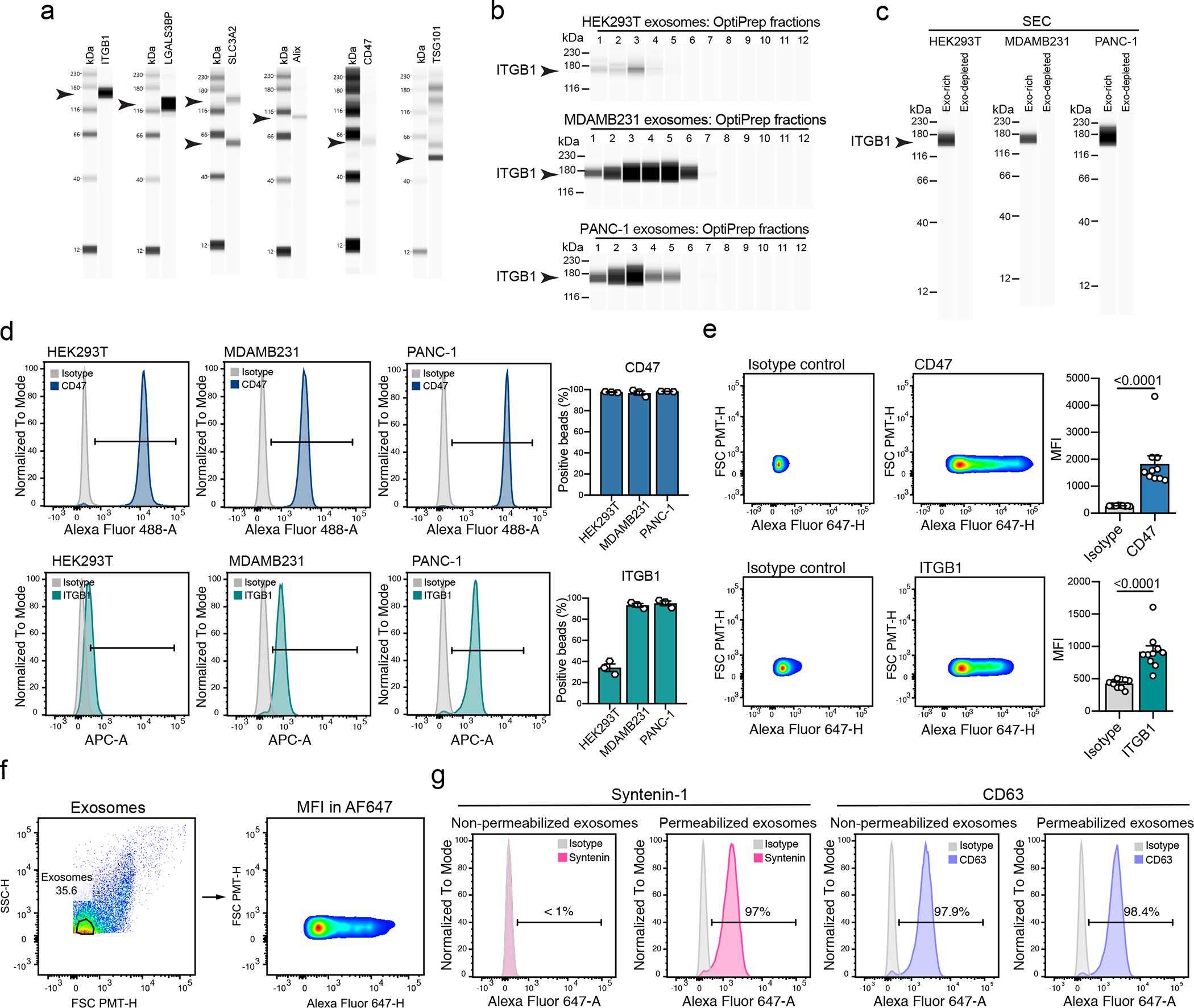
(a) Western blot analysis of ITGB1, LGALS3BP, SLC3A2, Alix, CD47 and TSG101 in UC-isolated exosomes from HEK293T cells. Results from one biological replicate. (b) Western blot analysis of ITGB1 in exosomes from the OptiPrep fractions of HEK293T, MDAMB231 and PANC-1. Results from one biological replicate. (c) Western blot analysis of ITGB1 in exosomes from the exosome-rich and exosome-depleted SEC fractions, from HEK293T, MDAMB231 and PANC-1. Representative results from three biological replicates. (d) Representative histograms show CD47 and ITGB1 levels in comparison to isotype control-stained beads in HEK293T, MDAMB231 and PANC-1-derived exosomes. Bar graph shows mean +/− s.e.m. of percentage of positive beads, individual data points from three biological replicates. (e) Representative histograms show the profile of the CD47 and ITGB1 levels in comparison to isotype control-stained exosomes from human plasma. Bar graph shows MFI +/− s.e.m, and individual data points refer to ten donors. Statistical significance was determined using two-tailed unpaired Mann Whitney t-test, and the exact p-values are shown. Significance defined as p < 0.05. (f) Gating strategy used for the FACS-based single particle analysis of exosomes. (g) Representative histograms show Syntenin-1 and CD63 positivity in non-permeabilized and permeabilized exosomes, in comparison to isotype control stained beads-bound exosomes.
