Fig. 3. The biochemical characteristics of amino acid residues within the V(D)J junctional portion of CDR3 calibrate the initial self-reactivity values of T cells.
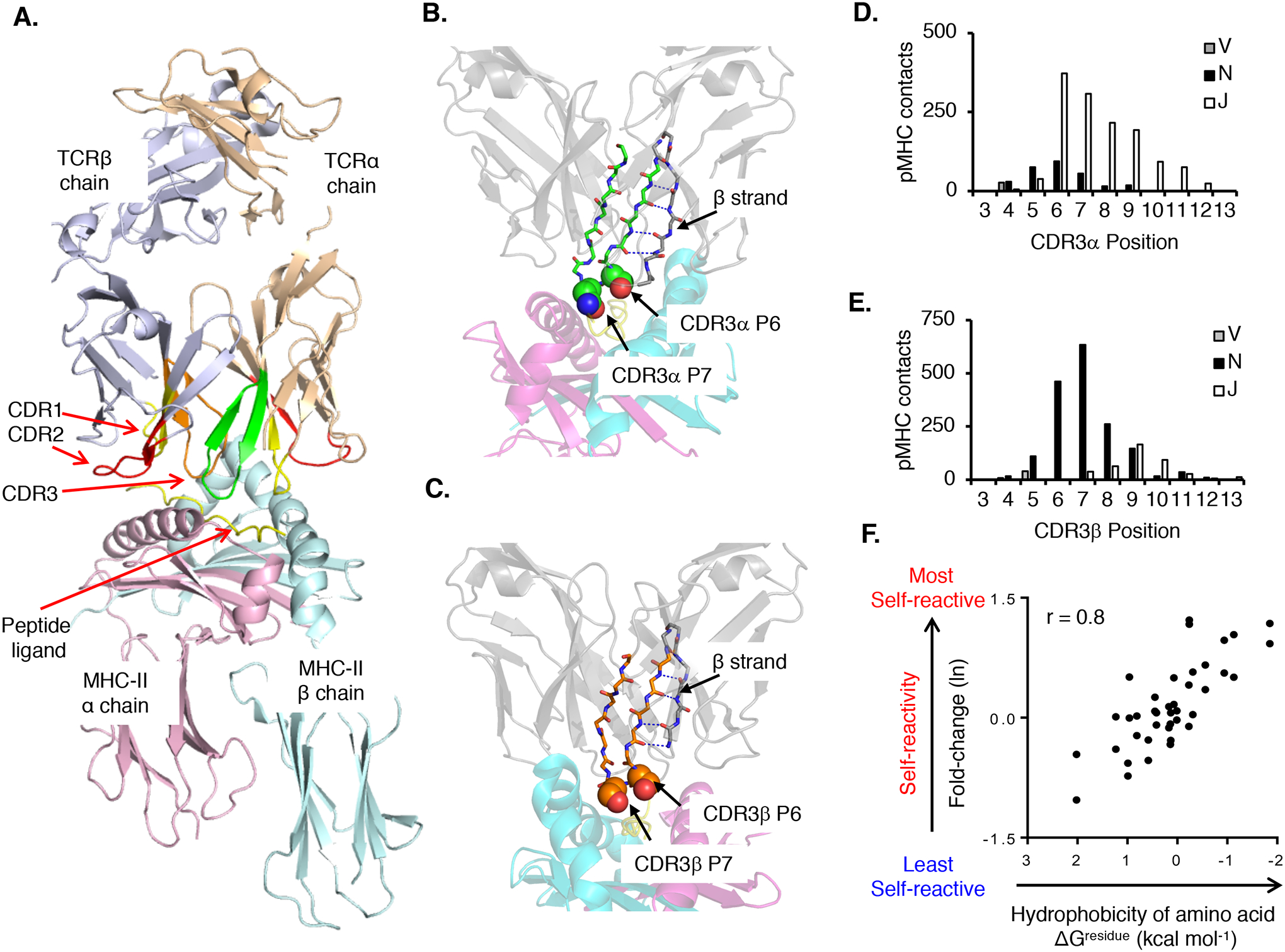
(A) Canonical orientation of an αβ TCR binding to pMHC. Highlighted are the TCRα chain (wheat) and TCRβ chain (purple), CDR1 (red), CDR2 (yellow), and CDR3α (green) and CDR3β (orange) binding to a pMHC-II ligand (PDB: 3C5Z). (B and C) A conserved β-strand within the TCRα (B) and the TCRβ V domains F-G loop (C) places CDR3 residues 6 (P6) and 7 (P7) at surface-exposed positions within the ligand-binding site. (D and E) CDR3α (D) and CDR3β (E) residues 6 and 7 form the most contacts with the pMHC. CDR3α residues contacting pMHC are primarily derived from Jα gene segments (~80%), whereas CDR3β residues contacting pMHC are primarily derived from N-region additions and Dβ gene segments (~80%). (F) Relative probability that a TCR will be self-pMHC reactive based on amino acid usage at CDR3 P6 or P7 as compared to the interfacial hydrophobicity value (ΔGresidue kcal mol−1) of each amino acid (98, 358). Each symbol represents an individual amino acid (n = 40). CDR3 contacts were assessed by using the following PDB accession codes: murine TCR–peptide–MHC class I: 1FO0, 1G6R, 1KJ2, 1LP9, 2CKB, 2O19, 2OL3, 3PQY, 3RGV, 3TF7, and 4MS8; murine TCR–peptide–MHC class II: 1D9K, 1U3H, 2PXY, 2Z31, 3C5Z, 3C6L, 3C60, 3QIB, 3QIU, 4P2Q, 4P2R, 4P5T, and 4P23; human TCR–peptide–MHC class I: 1AO7, 1BD2, 1MI5, 1OGA, 2AK4, 2BNQ, 2ESV, 2NX5, 3DXA, 3FFC, 3GSN, 3HG1, 3KPS, 3MV7, 3O4L, 3PWP, 3QDJ, 3SJV, 3UTS, 3UTT, and 4EUP; and human TCR–peptide–MHC class II: 1FTY, 1YMM, 1ZGL, 2IAM, 3O6F, 3PL6, 4E41, 4GRL, 4H1L, and 4P4K. Contacts were defined as two atoms localized within 4.0 Å, Ncont, CCP4 program suite 6.2.0.
