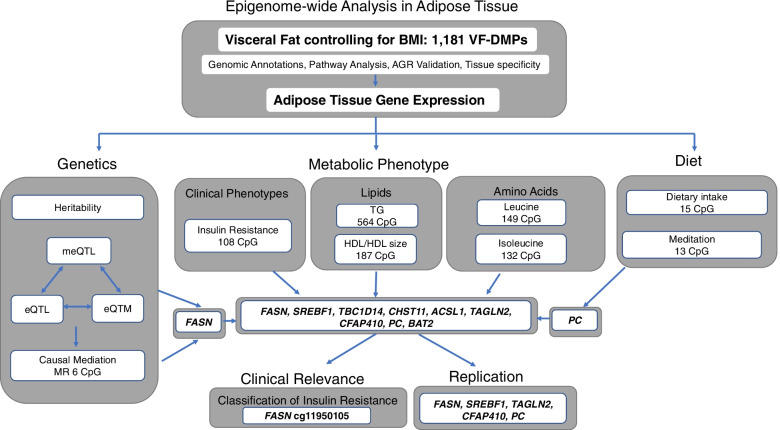
Fig. 1.
Study design. Epigenome-wide analyses of visceral fat (VF) were performed in 538 SAT samples, identifying 1181 VF-DMPs. The 1181 VF-DMPs were subject to downstream analyses assessing functional relevance using genomic annotations, tissue specificity, validation using a related phenotype android/gynoid ratio (AGR), and corresponding SAT gene expression changes related to VF. The VF-DMPs were then further analysed for association with genetic and lifestyle factors. Lastly, the signals were exploring using deep metabolic profiling across metabolic phenotypes, including lipid levels and metabolomic profiles, with replication. The resulting analyses were integrated to identify a set of replicated signals in genes with strong relevance to metabolic health