FIGURE 2.
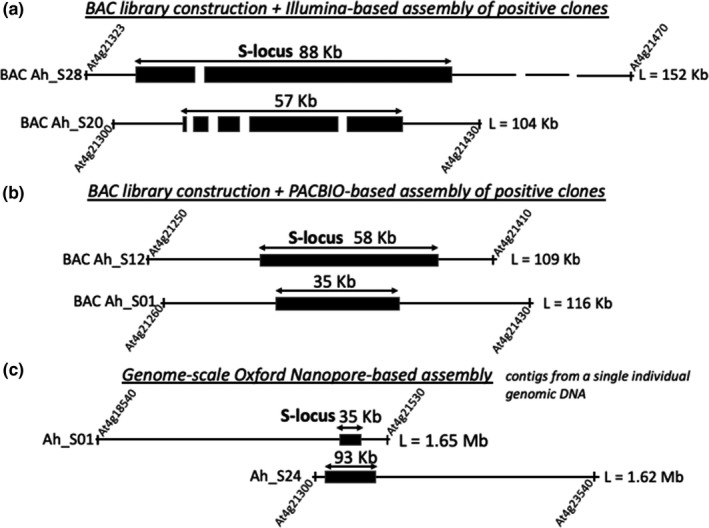
Influence of sequencing technology on level of assembly of individual haplotypes at the self‐incompatibility locus (S‐locus) in Arabidopsis halleri. (a) BAC clones sequenced using short‐read Illumina technology, showing fragmented assembly of the S‐locus despite starting from BAC clones (Goubet et al., 2012). (b) BAC clones sequenced using long‐read PacBio technology, showing complete assembly of the S‐locus after construction of BAC libraries and screening for the S‐locus region (V. Castric, unpublished data). (c) Whole‐genome sequencing using Oxford Nanopore technology, showing large contig sizes encompassing the full S‐locus for the two S‐haplotypes of a heterozygous individual (V. Castric, unpublished data). Limits of the contigs are annotated using A. thaliana gene IDs
