FIGURE 5.
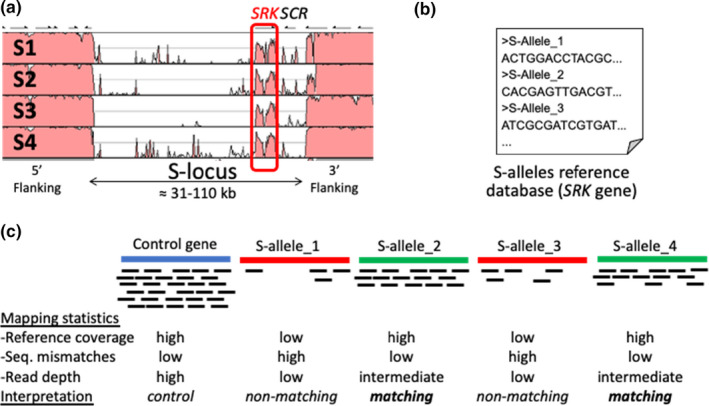
Strategy to infer S‐locus genotypes from individual short‐read resequencing data. (a) VISTA plot of haplotypes at the S‐locus of Arabidopsis halleri showing extremely low sequence conservation, except for the pistil‐expressed gene SRK (adapted from Goubet et al., 2012). (b) Schematic representation of a database of fasta sequences from previously known alleles at the SRK gene, which are used as references for sequential mapping of short‐reads from individual resequencing data. (c) Schematic representation of the results of sequential mapping against control genes (i.e., single‐copy genes with low polymorphism) or against nonmatching versus matching S‐alleles. The focal individual is heterozygous for S‐allele_2 and S‐allele_4, as mapping statistics against these two references are reporting high coverage of the reference sequence (proportion of positions with at least 1 read aligned), low sequence mismatches (between the reads consensus and the reference sequence), and intermediate (about half) read depth as compared to that for control genes (because of heterozygosity at the S‐locus)
