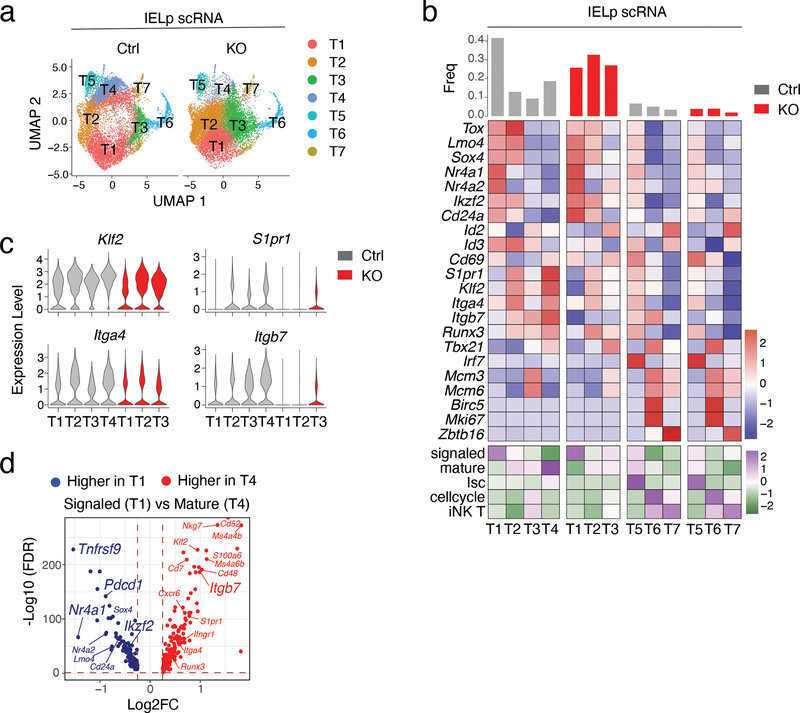
Figure 5. Impact of LRF on the IELp transcriptome.
(a-d) scRNAseq of thymic IELp.
(a) UMAP analysis of Ctrl and LRF KO IELp, displayed separately by genotype. Each dot is a cell and is color-coded by cluster (T1-T7), as defined in (b).
(b) Expression of selected genes (top, blue-red scale) and signature scores (bottom, green-purple scale) among IELp clusters defined on the integrated set of cells from both genotypes. The top bar graph indicates the frequency of cells in each cluster within the indicated genotype. Genotype-specific components of clusters T1-T4 (left two panels) and T5-T7 (right panels) are displayed separately. Gene expression values and signature scores are row z-scored on all clusters; color scales are indicated at the bottom right of each panel. Signaled, mature, Isc, cell-cycling, iNK T gene signature include genes with higher (FDR<0.05, Log2FC >0.25) expression in each of Ctrl clusters T1, T4, T5, T6, or T7, respectively, compared with all the other Ctrl cells.
(c) Violin plots show gene expression levels in selected clusters from indicated genotype.
(d) Volcano plot showing differentially expressed genes (FDR<0.05, |Log2FC| >0.25) between signaled (T1) and mature (T4) Ctrl IELp. The x-axis represents the mature over signaled average Log2 fold change (Log2FC); the y-axis shows -Log10 transformed FDR (adjusted p-value). Blue and red symbols indicate genes preferentially expressed in signaled and mature IELp, respectively. p-values were returned by Seurat FindMarkers function using a Wilcoxon Rank Sum test.