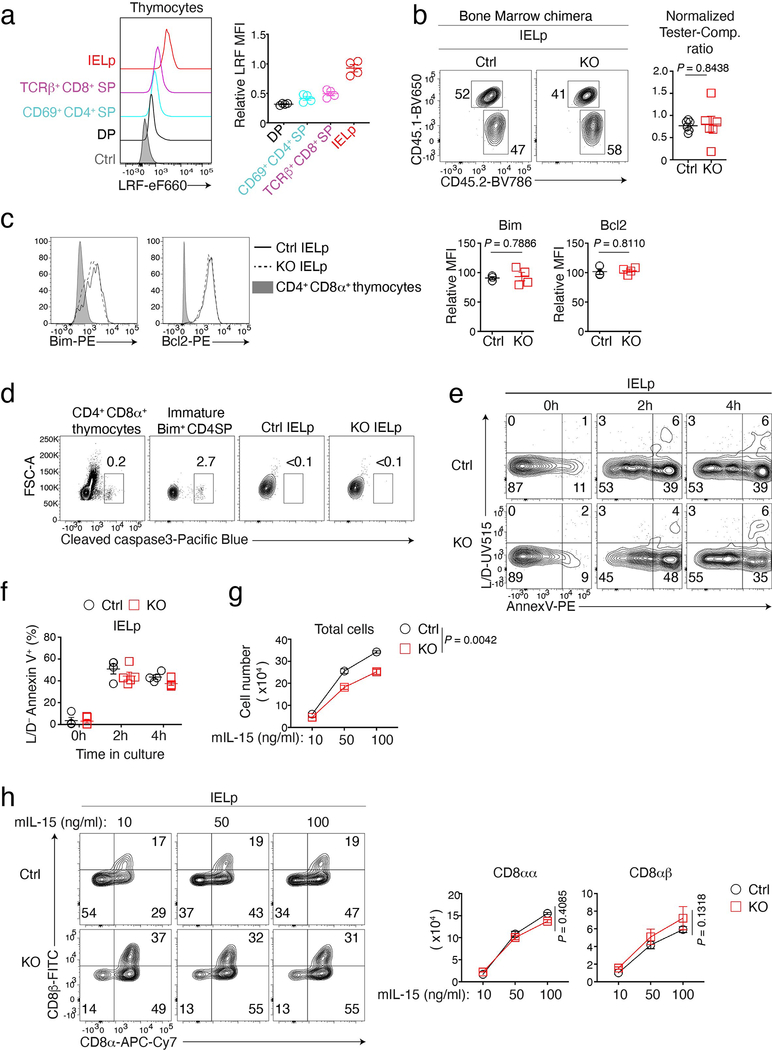
Extended Data Fig. 2. Development of LRF KO IELp.
(a) LRF expression in indicated subsets from Cd4cre+ Thpokfl/fl mice (to exclude cross reactive staining of Thpok by the LRF antibody). LRF KO TCRβ+ CD4−CD8+ splenocytes are shown as a control (grey-shaded). Data are from one experiment representative of two with 4 mice total. Graph (right) summarizes LRF expression (MFI) of indicated cells relative to that in wild-type IELp (analyzed as reference in each experiment), set to 1.
(b) CD45.2 vs. CD45.1 expression in IELp from bone marrow chimera analyzed in Fig. 1e. Graphs (right) show tester/competitor ratios in IELp normalized to tester/competitor ratio of B220+ splenocytes and summarize two independent experiments totaling 6 mice per group.
(c) Overlaid expression of Bim (left) and Bcl2 (right) in gated Ctrl and LRF KO IELp. Graphs (right) show the indicated protein expression (MFI) in IELp relative to that in IELp from wild type mice, set at 100 in each experiment. Data summarizes two independent experiments totaling 3 Ctrl and 4 LRF KO mice.
(d) Contour plots show cleaved Caspase3 levels vs. FSC in wild-type CD4+CD8α+ or immature Bim+ CD4+ SP (Bim+ CD4+ CD8− CD69+ MHC-I−) thymocytes, and in Ctrl and LRF KO IELp. Data are from one experiment representative of two with a total of 3 Ctrl mice and 4 LRF KO mice.
(e, f) Staining for extra-cellular annexin V and cell viability (L/D) on Ctrl and LRF KO IELp after in vitro culture for 0h, 2h or 4h (e). Graph (f) summarizes the percent of L/D−Annexin V+ cells among IELp, from 4 Ctrl and 5 LRF KO mice analyzed in two independent experiments.
(g, h) Total numbers of (g), and CD8α vs. CD8β expression by (h, left), Ctrl and LRF KO IELp after 4-day in vitro culture with the indicated IL-15 concentration. Right graph in (h) shows numbers of CD8αα and CD8αβ cells. Data is from three determinations for each genotype, acquired in two independent experiments
Error bars indicate standard error of the mean (SEM). P values are from two-tailed unpaired t-test (b, c) or two-way ANOVA (g, h). (a-c, f-h) Each symbol in graphs represents one mouse.