Figure 2.
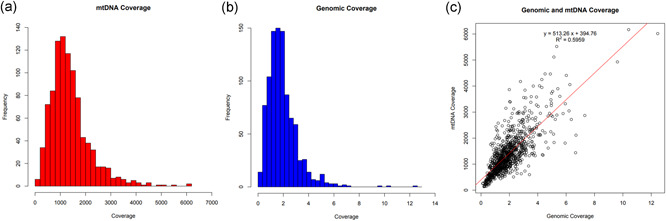
Comparison of sequencing coverage between the mitochondrial and nuclear genomes. (a) Mitochondrial genome (mtDNA) coverage for the low‐coverage whole‐genome sequencing (lcWGS) samples. The mean mtDNA read depth for the 957 samples that passed the haplogroup identification step was 1389X, with a standard deviation of 797.6X. The range of coverage was from 135.1X to 6169X. (b) Nuclear genome coverage for the lcWGS samples. The mean nuclear genome read depth for the 957 samples that passed the haplogroup identification step was 1.938X, with a standard deviation of 1.200. The range of coverage was from 0.1205X to 12.46X. (c) Comparison of the nuclear genome coverage to the mtDNA coverage across all samples. There was a moderate positive correlation between nuclear genome coverage and mtDNA coverage, with an R 2 value of .5959. Please note that the scale for the y axis (mtDNA coverage) is 500 times that of the x axis (nuclear genome coverage) to aid in the visualization of the line of best fit
