Figure 4.
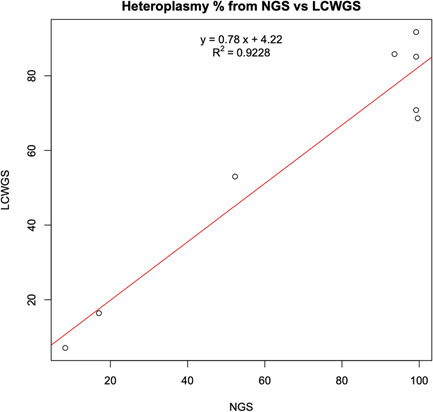
Correlation of estimated heteroplasmy between traditional PCR‐NGS and lcWGS. The heteroplasmy level for the eight pathogenic variants detected in the 960‐sample data set was calculated based on the novel lcWGS pipeline, versus a traditional PCR‐NGS methodology. The two methods showed a high degree of correlation for these variants, with an R 2 value of .9228. lcWGS, low‐coverage whole‐genome sequencing; NGS, next‐generation sequencing; PCR, polymerase chain reaction
