Table 1.
Simulation results of the proposed MMF and competing methods. Average errors in estimating 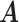 and
and 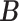 are quantified as the Hamming distance between the estimated and true matrices, normalized by the respective total number of elements. The numbers in the parentheses are standard deviations. The smallest errors are in boldface. The competing methods are low rank approximation (LRA), non-negative matrix factorization (NNMF), zero-inflated Poisson factor model (ZIPFM), and two-step multinomial matrix factorization (TSMF)
are quantified as the Hamming distance between the estimated and true matrices, normalized by the respective total number of elements. The numbers in the parentheses are standard deviations. The smallest errors are in boldface. The competing methods are low rank approximation (LRA), non-negative matrix factorization (NNMF), zero-inflated Poisson factor model (ZIPFM), and two-step multinomial matrix factorization (TSMF)
|
(2, 0.7) | (3, 0.6) | (5, 0.5) | |||
|---|---|---|---|---|---|---|
Error 
|
Error 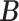
|
Error 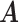
|
Error 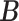
|
Error 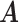
|
Error 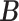
|
|
| MMF | 0.373 | 0.167 | 0.171 | 0.055 | 0.117 | 0.057 |
| (0.062) | (0.029) | (0.022) | (0.021) | (0.023) | (0.028) | |
| LRA | 0.298 | 0.269 | 0.205 | 0.185 | 0.203 | 0.165 |
| (0.042) | (0.023) | (0.052) | (0.017) | (0.058) | (0.021) | |
| NNMF | 0.351 | 0.279 | 0.288 | 0.247 | 0.256 | 0.208 |
| NNMF | (0.049) | (0.030) | (0.062) | (0.021) | (0.059) | (0.014) |
| ZIPFM | 0.425 | 0.258 | 0.291 | 0.249 | 0.246 | 0.232 |
| (0.014) | (0.031) | (0.008) | (0.023) | (0.002) | (0.022) | |
| TSMF | 0.382 | 0.253 | 0.325 | 0.132 | 0.237 | 0.089 |
| TSMF | (0.092) | (0.033) | (0.057) | (0.043) | (0.092) | (0.018) |
