Fig. 1.
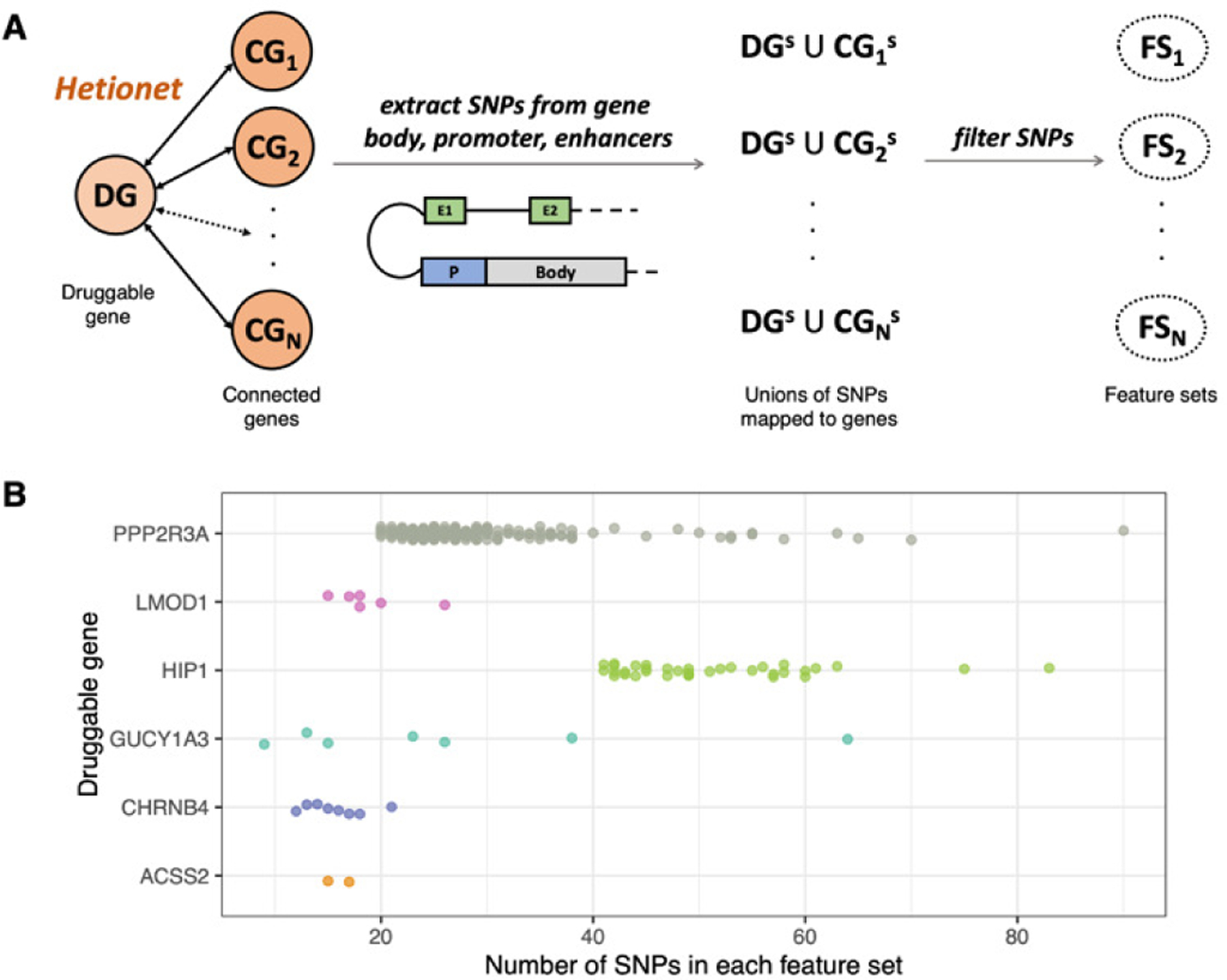
(A) Selection of SNPs and FSs. For each druggable gene DG, its connected genes (CGs) were obtained from Hetionet. For each CG, the SNPs mapping to its body, promoter, and putative enhancers were identified (CGS) and added to those mapping to DG (DGS). The corresponding FS was derived from these SNPs after filtering by functionality scorers and pruning. (B) FSs for the druggable genes. Each point corresponds to an FS for the DG indicated on the y-axis and its x-coordinate indicates the number of SNPs in that FS.
