Figure 3.
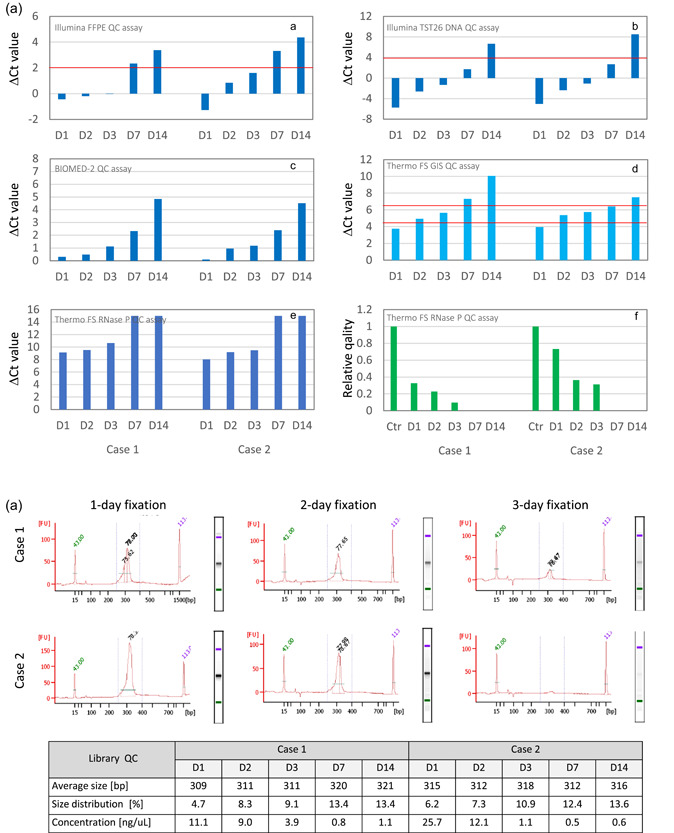
Effects of prolonged formalin fixation on DNA and next‐generation sequencing (NGS) library quality. The effects of formalin fixation time (1, 2, 3, 7, and 14 days) were examined at one institution using colorectal cancer specimens. DNA quality was determined by performing a real‐time PCR assay following DNA extraction using commercial formalin‐fixed paraffin‐embedded (FFPE) tissue DNA extraction kits (Qiagen). The libraries for amplicon sequencing (TruSeq Amplicon Cancer Panel; Illumina) were prepared using the MiSeq system (Illumina) and samples that passed the quality control (QC) assay. (a) The sample quality was determined by measuring the ΔC t values using the FFPE QC assay (Illumina) recommended for this gene panel. Samples fixed for 7 or 14 days did not pass the QC assay [A]. The other assay results were similar [B–F] (red line indicates cut‐off value). In the GI‐SCREEN study described above, the samples were classified into the three quality categories based on the results of assays performed using primer sets for specific housekeeping genes that differed in amplicon size [D]. As the FFPE blocks used in this analysis were prepared 3 years earlier, the ΔC t value of 2‐day‐fixed samples was classified as “intermediate.” For assays using primer sets for the RNase P gene that yield a different amplicon size, using an indicator utilizing calibration curves [F] instead of the usual ΔC t values [E] is recommended. (b) The libraries were prepared using 150 ng DNA, and library QC checks were performed on the Bioanalyzer 2100 (Agilent). Samples with 7‐ and 14‐day fixation that did not pass the ΔC t value‐based QC assay failed to produce library peaks. All samples with 1, 2, or 3‐day fixation produced library peaks and yielded sequencing results, although the peaks in the Case 2 sample with a 3‐day fixation were minute
