Figure 2:
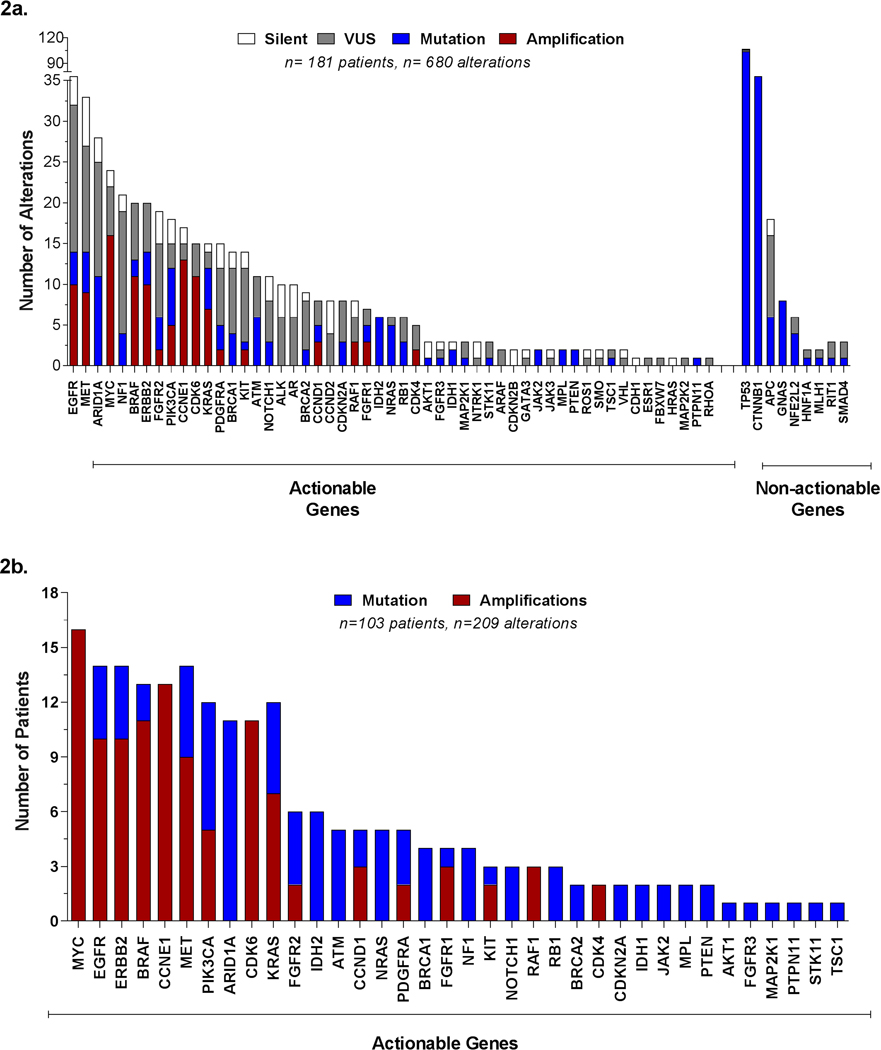
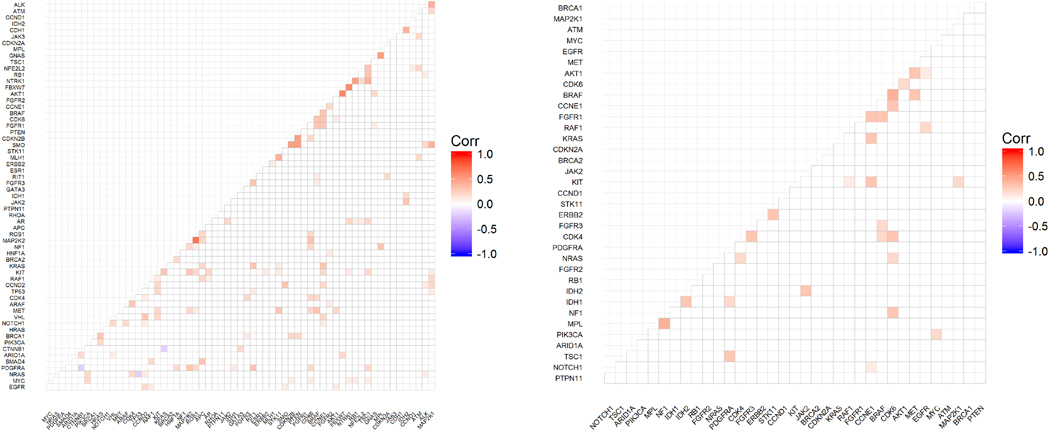
Most frequently mutated genes identified in cfDNA among HCC patients.
a) Distribution of all alterations identified, actionable and non-actionable genes.
b) Proportion of patients with alterations in actionable genes only.
c) Gene correlation matrix among all patients with alterations detected. All patients with alterations detected, irrespective of the actionability of the alteration (i.e. variant in non-actionable genes, VUS, and/or synonymous) were considered for this matrix. In all from 181 patients and 62 genes were evaluated with Spearman’s rank correlation and displayed in the correlation matrix above. Non-significant correlations are marked with “blank” in the graph. Values < 0 indicate no agreement, 0 – 0.20 slight agreement, 0.21 – 0.40 fair agreement, 0.41 – 0.60 as moderate agreement, 0.61 – 0.80 as substantial agreement, and 0.81 – 1 as almost perfect agreement.
d) Gene correlation matrix among HCC patients with alterations in actionable genes (synonymous and VUS alteration excluded). Exclusion of non-actionable genes, VUS, and synonymous alterations reduced the patient population size considered for this matrix from 181 to 103 patients and from 62 genes to only 36 actionable genes. The correlation between mutated genes was evaluated with Spearman’s rank correlation and displayed in the correlation matrix above. Non-significant correlations are marked with “blank” in the graph. Values < 0 indicate no agreement, 0 – 0.20 slight agreement, 0.21 – 0.40 fair agreement, 0.41 – 0.60 as moderate agreement, 0.61 – 0.80 as substantial agreement, and 0.81 – 1 as almost perfect agreement.
