SUMMARY
Eremophila is the largest genus in the plant tribe Myoporeae (Scrophulariaceae) and exhibits incredible morphological diversity across the Australian continent. The Australian Aboriginal Peoples recognize many Eremophila species as important sources of traditional medicine, the most frequently used plant parts being the leaves. Recent phylogenetic studies have revealed complex evolutionary relationships between Eremophila and related genera in the tribe. Unique and structurally diverse metabolites, particularly diterpenoids, are also a feature of plants in this group. To assess the full dimension of the chemical space of the tribe Myoporeae, we investigated the metabolite diversity in a chemo‐evolutionary framework applying a combination of molecular phylogenetic and state‐of‐the‐art computational metabolomics tools to build a dataset involving leaf samples from a total of 291 specimens of Eremophila and allied genera. The chemo‐evolutionary relationships are expounded into a systematic context by integration of information about leaf morphology (resin and hairiness), environmental factors (pollination and geographical distribution), and medicinal properties (traditional medicinal uses and antibacterial studies), augmenting our understanding of complex interactions in biological systems.
Keywords: Eremophila, Scrophulariaceae, metabolomics, Myoporeae, phylogeny, diterpenoid, serrulatane, viscidane
Significance Statement
The chemical space of the Australian genus Eremophila, used in traditional medicine by Indigenous Australians, is explored using a novel large‐scale computational metabolomics‐based molecular network approach combined with phylogenetic analyses to provide a detailed overview of the complex metabolite diversity and chemo‐evolutionary relationships present. Integration of information about leaf morphology (resin and hairiness), environmental factors (pollination and biogeographical distribution), and medicinal properties (Aboriginal uses and antibacterial studies) brings the chemical information into a systematic context.
INTRODUCTION
The large, cosmopolitan plant family Scrophulariaceae sensu stricto contains eight tribes including the Myoporeae. Myoporeae contains seven genera, with Eremophila being the largest genus in the tribe (approximately 230 species and 58 subspecies). Eremophila exhibits incredible morphological diversity throughout the Eremean (arid) biome, which covers approximately 70% of the Australian continent. Indigenous people of the Australian mainland – the Australian Aboriginal Peoples – recognize Eremophila species as important sources of traditional medicine (Chinnock, 2007; Richmond, 1993; Richmond and Ghisalberti, 1994). Aboriginal Peoples use a range of Eremophila species and preparation methods, though use of leaves is documented most commonly. Ethnomedicine can be a key guide to bioactive natural products and potential drug leads (Gyllenhaal et al., 2012; Ngo et al., 2013). Chemical exploration of Eremophila and closely related species was pioneered by Emilio Ghisalberti and colleagues in the 1970s (Ghisalberti et al., 1975). A plethora of unique and structurally diverse metabolites belonging to the family of terpenoids have subsequently been uncovered (Biva et al., 2016; Gericke et al., 2020; Kjaerulff et al., 2020; Ndi et al., 2007a; Pedersen et al., 2020; Sadgrove et al., 2021; Singab et al., 2013; Tahtah et al., 2016; Wubshet et al., 2016) (Figure 1). Extracts from Eremophila species have been tested for antiviral (Semple et al., 1998), antibacterial (Ndi et al., 2007b; Palombo and Semple, 2001, 2002), and anticancer (Mijajlovic et al., 2006) activity, and show inhibitory activity of ion channels (Rogers et al., 2002). Isolated serrulatane‐ and viscidane‐type diterpenoids exhibit antimalarial (Kumar et al., 2018), antibacterial (Algreiby et al., 2018; Barnes et al., 2013; Biva et al., 2019; Liu et al., 2006; Mon et al., 2015; Ndi et al., 2007a, 2007c), antidiabetic (Tahtah et al., 2016; Wubshet et al., 2016), and antiinflammatory (Liu et al., 2006; Mon et al., 2015) properties. The presence and biosynthesis of interesting metabolites in species of Myoporeae have previously been demonstrated (Gericke et al., 2020; Kjaerulff et al., 2020; Kracht et al., 2017; Kumar et al., 2018; Ndi et al., 2007b; Pedersen et al., 2020; Tahtah et al., 2016; Wubshet et al., 2016) and recent phylogenetic studies have revealed complex evolutionary relationships between genera in the tribe (Fowler et al., 2020, 2021). Still only a fraction of species within the Myoporeae have had their metabolites investigated. Based on the rich source of bioactive molecules this group has proven to be, it is expected that further investigation will yield many more novel molecules with potential as pharmaceuticals.
Figure 1.
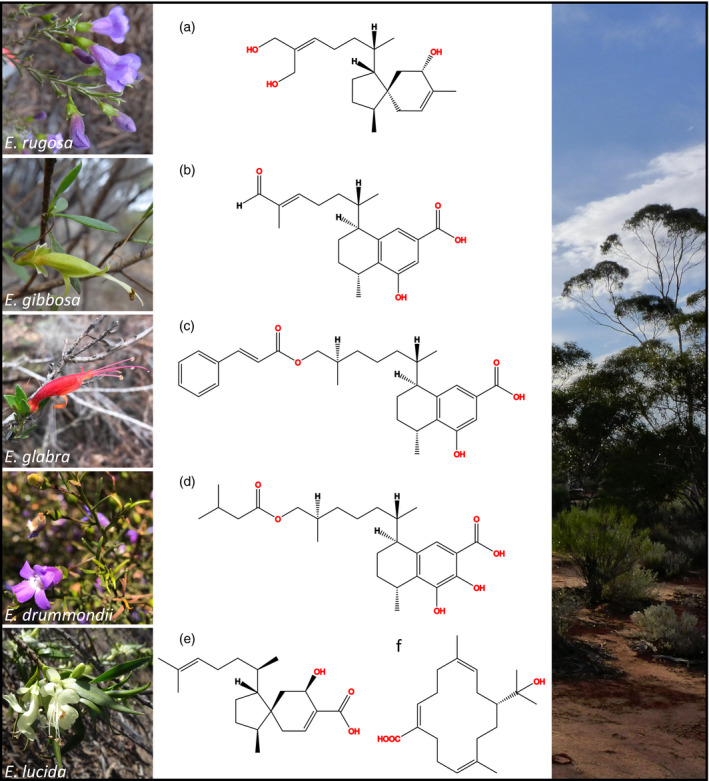
Chemical and morphological representation of selected Eremophila species.
Images on the left display selected Eremophila species. Corresponding diterpenoids isolated in prior studies are displayed in the middle column. The right column shows a typical Eremean habitat in Western Australia. (a) Eremophila rugosa, KU036‐6‐5 (Zhao et al., unpublished data). (b) Eremophila gibbosa, 8‐hydroxy‐16‐oxoserrulat‐14‐en‐19‐oic acid (Wubshet et al., 2016). (c) Eremophila glabra, 8‐hydroxy‐16‐cinnamoyloxyserrulat‐19‐oic acid (Wubshet et al., 2016). (d) Eremophila. drummondii, 7,8‐dihydroxy‐16‐[3‐methylbutanoyloxy]serrulat19‐oic acid (Wubshet et al., 2016). (e, f) Eremophila lucida, (e) 5‐hydroxyviscida‐3,14‐dien‐20‐oic acid and (f) (3Z, 7E, 11Z)‐15‐hydroxycembra‐3,7,11‐trien‐19‐oic acid (Tahtah et al., 2016).
Plant diterpenoids represent a rich source of bio‐based pharmaceuticals and constitute the foundation of many drug development success stories (Mafu and Zerbe, 2018), such as paclitaxel (Taxol) (Engels et al., 2008), ingenol mebutate (Picato) (Ogbourne and Parsons, 2014), and forskolin (e.g., ForsLean) (Pateraki et al., 2017). Despite evidence of diterpenoids as high‐value compounds, only a small fraction of plant species have been thoroughly investigated in drug discovery campaigns (Harvey et al., 2015; Sharma and Sarkar, 2013). The hunt for novel plant‐derived pharmaceuticals is not a trivial task given the immense number of unknown metabolites present in any given species. Emerging tools that can help in this challenge are in silico dereplication methodologies that can illuminate this chemical ‘dark matter’ found in plants (da Silva et al., 2015). One such tool is molecular networking, an approach which greatly enhances the chemical annotation of metabolomics data and allows a streamlined hypothesis‐driven targeting of metabolites in contrast to the traditional ‘grind and find’ model (Aron et al., 2020; Fox Ramos et al., 2019; Kang et al., 2019; Wang et al., 2016). Molecular networking further enables integration of functional annotations, such as biological, taxonomic, and geographical data (Ernst et al., 2019b; Olivon et al., 2017, 2018). This multilayered approach facilitates the dereplication of large metabolite datasets associated with interesting functionalities and thus improves our understanding of biological systems in which they are found.
In this study, we have investigated the chemo‐evolutionary relationships in the plant tribe Myoporeae by applying phylogenetics and state‐of‐the‐art computational metabolomics to a dataset of 291 leaf samples of Eremophila and allied genera. Information about leaf morphology (resin and hairiness), environmental factors (pollination and geographical distribution), and medicinal properties (traditional medicinal uses and antibacterial studies) is combined with natural product chemistry, qualified by use of a large spectral dataset of 76 reference compounds. This allows for plant chemistry to be considered in a systematic context that augments our understanding of interactions in biological systems and facilitates targeted drug discovery. This research has been conducted under Australia’s access and benefit‐sharing laws which are consistent with obligations under the Nagoya Protocol (Secretariat of the Convention on Biological Diversity, 2011).
RESULTS AND DISCUSSION
Chemo‐evolutionary relationships in Myoporeae
In this study, molecular phylogenetics using data from high‐throughput DNA sequencing was integrated with plant metabolomics utilizing state‐of‐the‐art molecular networking tools to elucidate chemo‐evolutionary relationships by exploring metabolite diversity and evolution across the diverse tribe Myoporeae (Figure S1). Our uniquely comprehensive sampling included 291 specimens, representing six genera and approximately 80% of the species in the tribe (Table S1). We assessed metabolite diversity in Myoporeae using untargeted high‐resolution mass spectrometry (MS1 and MS2) to generate a molecular network via a feature‐based molecular networking pipeline (Nothias et al., 2020). This approach relies on the modified cosine score‐based matching of MS2 fragments (combines fragment m/z and mass shift by the precursor mass difference) (Wang et al., 2016), and thereby spectral features will be structurally assigned to each other based on set thresholds, such as the cosine score (here 0.8) and the minimum number of matched fragments (here eight). Cosine score‐based matching of spectral features is the most widely used method in current untargeted metabolomics studies, providing an exhaustive structural comparison of large spectral datasets. In general, cosine score‐based methods efficiently reveal similar spectra, but are less suited to handle highly modified molecules. Furthermore, spectral networking and library matching for dereplication strongly depend on the chosen parameters and algorithms used, and thus need to be adjusted carefully. Spectral features that do not match these networking requirements remain as singletons and do not partake in downstream spectral cluster‐based tools such as Network Annotation Propagation (NAP) (Da Silva et al., 2018). Often these singletons represent MS2 spectra of poor quality but also molecules with low level of fragmentation under the given analytical method. An adaptation to the cosine score‐associated limitations is the integration of neutral loss comparisons (Guijas et al., 2018; Moorthy et al., 2017). A promising new implementation of this method uses the concept of hypothetical neutral loss, which is calculated as the mass difference between a pair of fragment ions and contains core structural information that facilitates spectral similarity matching involving highly decorated compounds (Xing et al., 2020). A new direction in spectral similarity scoring was recently presented as ‘spec2vec’, a machine learning‐inspired spectral similarity scoring tool. Spec2vec learns meaningful relations between fragments and neutral losses in MS2 fragmentation spectra to better resemble the structural similarity of fragmented molecules, outperforming the cosine score, and thus serves as a suitable alternative method of evaluating spectral similarity (Huber et al., 2021).
To focus on the chemically diversified fraction of the Myoporeae metabolome, we used the 100 largest chemical subnetworks with at least nine nodes, each chemical subnetwork representing a chemical family of structurally related features (Figure 2). Those 100 chemical families altogether comprised 3329 nodes after normalization, representing 54% of the full connected network (6110 nodes), excluding singletons (4586 nodes without a structurally similar neighbor node). Within this dataset, the median number of detected chemical families per specimen was 15 and the maximum was 36. Singleton features present 43% of the studied chemical space and thus further investigation of the nature of these features was conducted. We found that most singletons are only found in one specimen (60%), while 18% and 8% are localized to two and three specimens, respectively. Based on the maximum signal intensity for each detected singleton, we found the median intensity to be 31 244, close to the intensity threshold (10 000), suggesting poor spectral quality as the underlying factor for these features to be singletons (Figure S2). For a more detailed examination, 10 features with the highest detected signal intensity were selected for a networking‐independent in silico assisted structural prediction using the software sirius (Dührkop et al., 2019). Thereby, we could predict three flavonoids, one sesquiterpenoid‐lactone, and five linear structures (one diterpenoid and four fatty acids), as well as one still unknown compound (Table S2). This result shows that there is indeed meaningful chemical information present within the singleton features. However, most singletons are rather sample‐specific features with overall very low abundance associated with poor spectral quality and thus not suitable for a molecular networking approach.
Figure 2.
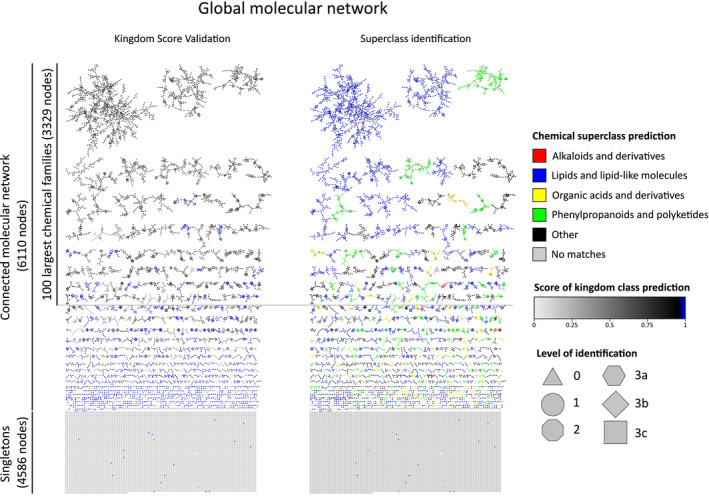
The global molecular network of Myoporeae.
The global molecular network of Myoporeae comprises a connected part, with clusters of spectral features that represent structurally related putative metabolites, as well as singletons, which represent features that did not show spectral similarity to others. The connected dataset was dereplicated using public, in‐house, and in silico spectral libraries and subsequent Network Annotation Propagation (NAP). Identification of nodes took place on six different levels, with level 1: m/z, retention time, and MS2 match using an in‐house spectral library; level 2: MS2 match using in‐house and public spectral libraries; level 3a–c: Fusion, Consensus, or MetFrag algorithm‐based in silico fragmentation database match; level 0: no spectral match. The presented network on the left displays a kingdom score‐based assessment with overall high scoring values that support a robust dereplication of the corresponding chemical families. The network on the right side displays NAP‐generated chemical superclass prediction for each chemical family, with selected classes highlighted in color.
Within our dereplication efforts, we were able to validate the presence of 37 compounds from our in‐house database containing 76 unique reference compounds previously identified in Myoporeae species (level 1 identification) (Table S3). The lack of 39 compounds is most likely due to the different origin and treatment (dried versus fresh) of the plant material used in this study. However, we found 11 reference compounds (30%) to be localized among the selected 100 chemical families, comprising serrulatane and viscidane diterpenoids, flavonoids, and verbascoside, thus representing the major chemical constituents found in Eremophila species (Singab et al., 2013).
Clear phylogenetic patterns, with closely related species having similar chemical profiles, were revealed when metabolic clusters and phylogenetic analyses were compared in the form of a tanglegram (Figure 3). A tanglegram is used to directly compare dendrograms, while an untanglement algorithm reveals similar clusters of specimens on both sides, which in this study displays chemo‐evolutionary relationships (Galili, 2015). Hereby, an entanglement factor evaluates the similarity of the compared dendrograms with ‘0’ displaying perfect congruency and ‘1’ a full mismatch. Chemical similarity among specimens was assessed based on binary presence/absence information of chemical families and using hierarchical cluster analysis. Here we decided on a metabolite inclusion threshold of ‘1’, which means that a specimen holds a chemical family if at least one respective spectral feature could be detected in its extract above a peak signal intensity of 10 000. Tanglegram analyses based on increased inclusion factors (2, 3, and 5) were generated and display a higher level of entanglement and a lower degree of connections of equal clustering with increasing inclusion thresholds (Figure S3e–g, Table S4). While we can still detect tanglegram metabolic cluster (TMC) A and B with an inclusion factor of ‘2’, this pattern fades when raising it further, which is also visualized within a heatmap representing the total number of features present of each chemical family (Figure S4). A fast metabolic flux that does not accumulate many compounds of a given chemical family as well as unique modifications of the same structural backbone in different specimens could be the underlying reasons for this frequently observed low abundance of features from the same chemical family. This further underlines the importance of describing this highly diversified chemical space to obtain insight into the chemical similarity between Myoporeae specimens. The binary assignment of spectral features facilitates the interpretation of large and diverse spectral datasets. While maintaining spectral quality by applying a reasonable threshold, in some cases spectral features could still fall just below it and thus will be falsely assigned as absent. Since chemo‐evolutionary relationships comprise many different features, these should not be affected by these individual cases. Nevertheless, we advise a final more detailed assessment of the spectral data of interest after applying the presented processing pipeline.
Figure 3.
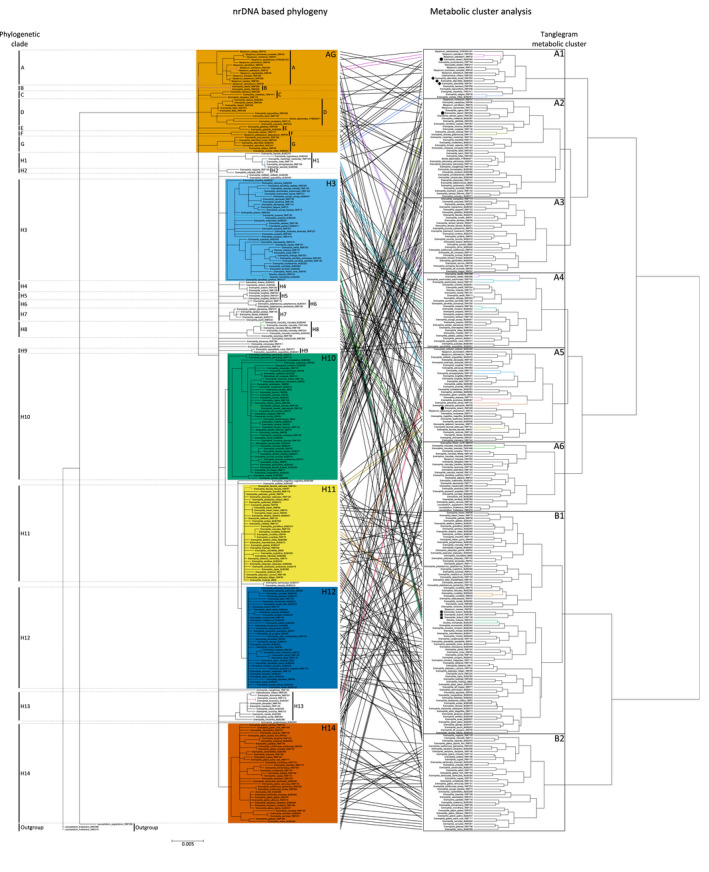
Molecular network‐based tanglegram analysis showing chemo‐evolutionary relationships in Myoporeae.
Tanglegram representation of conjoined phylogenetic and metabolic information from 291 specimens of tribe Myoporeae. The left side displays the Bayesian inference nuclear ribosomal DNA phylogeny. All bifurcating branches have strong support (posterior probability ≥ 0.95%); all nodes with support < 0.95 have been collapsed. All phylogenetic clades are labeled (clades A–H), and major ones are highlighted by color code. The right side displays the metabolic cluster analysis that was conducted based on the presence or absence of the 100 largest chemical families within the generated molecular network of Myoporeae. Therein, tanglegram metabolic clusters (TMCs) are indicated. The tanglegram analysis connects the same specimens by a line, which is colored when equal clustering of specimens is present in both analyses. Selected species with varying interspecific chemical variation are highlighted within the metabolic cluster analysis, with square indicating E. duttonii, diamond indicating E. alternifolia, and circle indicating E. deserti.
The phylogenetic analysis identified eight major lineages (labeled phylogenetic clades A–H) and the metabolic cluster analysis identified two distinct TMCs, A and B (Figure 3). This clustering is evident in metabolic cluster analyses based on 100 chemical families as well as the full metabolite dataset containing the isolated 10 696 individual spectral features (Figure S3a), giving rise to similar chemo‐evolutionary patterns observed in respective tanglegrams. Thereby, the tanglegram based on 100 chemical families is slightly more entangled (entanglement factor 0.24 versus 0.19) and has a different degree of detected groups of equally clustered specimens (13 versus 20) (Table S4). Both tanglegrams display a similar structure, in particular within TMC B, while the separation of TMC A into A1–3 and A4–6 becomes more evident when applying the entire spectral dataset. This underlines that the selected 100 chemical families harbor the overarching chemo‐evolutionary information within the Myoporeae metabolome, while neglected spectral features are still meaningful. The latter is also reflected when analyzing a tanglegram based on the detected 4586 singletons, which still expresses some chemo‐evolutionary trends mostly in members of clades H14 and H10, indicating remaining information within these spectral features (Figure S3d).
TMC A includes the majority of Eremophila diversity, all allied genera, and both outgroup species (Leucophyllum) (Table 1). Morphologically, species producing metabolic features characteristic of TMC A are highly diverse, being variously insect‐ or bird‐pollinated and having hairy or glabrous and resinous or non‐resinous leaves. Geographically, species of TMC A are represented across the full distribution of the tribe, throughout Australia’s arid and semi‐arid zones, but also including temperate, subtropical, and non‐Australian members. Notably, the majority of species found in TMC A1 correspond to species placed in phylogenetic clades A–G, which are considered to be potentially early diverging lineages of the tribe, sister to phylogenetic clade H. From a biogeographic perspective, species in TMC A1 exhibit a distinctly non‐arid distribution that sets them apart from all other clusters. In contrast, species harboring features characterizing TMC B are a subset of Eremophila species from three main lineages of phylogenetic clade H (subclades H11, H12, and H14). Species sharing the chemical characteristics of TMC B are also morphologically diverse (bird‐ and insect‐pollinated, hairy and glabrous leaves) and are more likely to have some particular traits: within the TMC B taxa, 83% of the species have resinous leaves whereas this only applies to 31% of the TMC A taxa. Species in TMC B occur largely in Australia’s central arid zone, with particularly high species diversity in the Northwest (TMC B1) and Southwest (TMC B2) of Western Australia relative to species associated with TMC A chemistry (Figure S5).
Table 1.
Summary of morphological, environmental, and medicinal attributes between major tanglegram metabolic clusters (TMCs). Attributes are evaluated between the two dominant TMCs, A and B, which are based on the presence/absence of the 100 largest chemical families found within the investigated Myoporeae‐associated molecular network
| TMC A | TMC B | |
|---|---|---|
| Total no. of species | 130 (+ 2 outgroup species) | 85 |
| Genera represented, no. of species in brackets | Bontia (1) | Diocirea (2) |
| Calamphoreus (1) | Eremophila (82) | |
| Diocirea (1) | Myoporum (1) | |
| Glycocystis (1) | ||
| Eremophila (114) | ||
| Myoporum (12) | ||
| No. of resinous:non‐resinous taxa (duplicate specimens removed) | 45:99 (31% resinous:69% non‐resinous) | 86:18 (83% resinous:17% non‐resinous) |
| No. of taxa with hairy leaves:glabrous leaves | 90 hairy:54 glabrous (62%:38%) | 76 hairy:28 glabrous (73%:27%) |
| No. of insect‐:bird‐pollinated species (duplicate specimens removed) | 111:36 (75% insect:25% bird) | 67:37 (64% insect:36% bird) |
| Documented traditional use | 10 species | 8 species |
| Documented antimicrobial activity | 8 species (of total 51 taxa tested [8 + 43]) | 25 species (of total 37 taxa tested [25 + 12]) |
Plants are known to express a highly versatile chemistry throughout time and space (Kessler and Kalske, 2018). This metabolic variability should be considered when plants are sampled for chemical analysis; such data display a chemical snapshot from a sample at a given state of plant ontogeny and influenced by growth conditions and environmental challenges and time. Although metabolite modification and concentration may fluctuate drastically between individuals of the same species, we suggest that plants with a similar metabolome contain the same structural scaffolds, and thus basal chemical relationships can still be deduced. To assess and account for such variation, our dataset intentionally contains duplicate collections for over 50 species, as many species exhibit considerable morphological variation and wide geographic distributions that in some cases span thousands of kilometers. For some species that are supported as monophyletic, an overarching similarity in their metabolic profiles was observed. For example, Eremophila alternifolia (TMC A1) and Eremophila duttonii (TMC B1) are extremely widespread in distribution (Figure S6) and yet despite spatial and temporal variation in specimen collection (collections were made at different times of day and year), each species forms a unique chemical cluster in the metabolic cluster analysis seen in Figure 3. In other instances the converse of the above scenario is observed, whereby well‐supported monophyletic species were found to be chemically quite diverse as seen in placement of individuals of the same species in separate TMCs. A good example of this can be seen in the examined specimens of Eremophila deserti, another widespread species with considerable within‐species morphological variation. Each of the three E. deserti specimens are placed in a different TMC (A1, A2, and A5), indicating considerable chemical variation within the species (Figure 3). By tracing the corresponding metabolite profiles among the global molecular network of Myoporeae, we observed commonly shared features as well as individual clusters of structurally related features for each specimen (Figure S7). This characterized E. deserti specimen MJB2384 as having an altered contingent of flavones (chemical families 17, 34, and 52), whereas E. deserti specimen RMF204 had a terpenoid‐enriched chemical profile with sesquiterpenes grouped into chemical family 7 and monoterpenes into chemical family 47 (Figure S8). In contrast, the metabolite diversity of E. deserti specimen RMF228 showed a rather wide contingent of features touching multiple chemical families of different classes, including terpenes and flavones. The distinct localization of E. deserti specimens within separated TMCs compared to E. alternifolia and E. duttonii specimens was also detected in metabolic cluster analyses conducted on the full metabolite dataset based on binary assignment as well as absolute peak signal intensity (Figure S3a,c). This highlights the need for further work into the main drivers of such intraspecific chemical variation, e.g., by analysis of transcriptomes from different stages of plant ontogeny or following abiotic or biotic stresses, or looking for biogeographic genetic structuring within a species. Nevertheless, focusing the observed chemical space towards families of structurally related metabolites within a molecular network allowed us to mitigate these effects in the presented metabolomic analysis, while capturing important relevant relationships among a large set of species.
Heatmap analyses integrating the observed chemical space with phylogenetic relationships in Myoporeae were used to explore possible associations between plant chemistry, resin production, and geography. This approach shows a clear phylogenetic signal, with specific groups of chemical families (heatmap metabolic clusters [HMCs]) found to be associated with phylogenetic clades (Figure 4). HMCs V, VIII, IX, and X show strong phylogenetic signatures in the heatmap, and the dereplication approach revealed a dominance of terpenoid‐related chemistry in these clusters, typically associated with serrulatane and/or viscidane diterpenoids. The complete lack of these chemical families in outgroup species, despite the dominance amongst many clades of Eremophila suggests that this chemistry may be unique and highly specialized within Myoporeae. Specifically, HMC IX chemistry is dominant in phylogenetic subclades H11 and H12 and is largely absent from other lineages. While also dominant in phylogenetic clades H11 and H12, HMC VIII chemistry extends to early diverging lineages (clades A–G) and HMC X extends to clade H14. Interestingly, HMC V is largely unique to phylogenetic clade H14.
Figure 4.

Categorial heatmap analysis displays the chemo‐evolutionary framework of Myoporeae.
A heatmap analysis was used to evaluate chemical information in an evolutionary context. Information from the 100 largest chemical families was compiled, which was derived from the molecular network of Myoporeae. For each specimen in this study, the presence (black) or absence (white) of at least one putative metabolite from a corresponding chemical family is displayed within the heatmap. Left: The nuclear ribosomal DNA phylogeny on the left side presents the major phylogenetic clades. Top: As depicted on top of the heatmap, the chemical information underwent hierarchical clustering according to the given phylogeny to reveal chemo‐evolutionary patterns among subsets of chemical families, defined as heatmap metabolic clusters (HMCs) I–X. Bottom: Selected information about chemical identity comprising different levels of classification, i.e., superclass and class, as well as annotations on the metabolite level (serrulatane‐ and viscidane‐type diterpenoids). Corresponding dereplication is indicated in blue, while the additional presence of level 1 identification (m/z, retention time, and MS2 match) in a particular chemical family is highlighted in gold. Right: Functional annotations including the presence of leaf resin and hairiness, pollination, antibacterial activity, and traditional usage, as well as biogeographical species distribution information, are displayed on the right side. A summary of legends is included to the right of the figure.
Resin chemistry as a key strategy to adapt to the Eremean zone
To relate chemo‐evolutionary relationships to specific plant and environmental characteristics, as well as to biological activity, we enhanced the heatmap analysis with metadata on leaf resin and hairiness, pollinator types, geographic distributions, traditional medicinal usage, and reported activities against human Gram‐positive bacterial pathogens (Table S1). This revealed a strong association between HMC IX and X chemistry and the formation and accumulation of leaf resin in phylogenetic clades H11–H14 (Figure 4). Independent generalized linear models for each HMC–metadata combination showed that resinous leaves are significantly associated with both HMC IX (manyglm, P = 0.001) and X (manyglm, P = 0.001) (Table S5). The species in these two HMCs directly correspond to the main members of TMC B with a high frequency of leaf resin relative to TMC A. We hypothesize that HMC IX and X chemistry is a main driver of tanglegram clustering patterns, based on its predominant presence in TMC B (Table S6). The abovementioned general linear model analysis revealed an overall highly significant influence of the presence of leaf resin towards individual HMCs. A global permutational multivariate analysis of variance (permanova) analysis supports this strong overall association between leaf resin and chemical diversity in the dataset, with leaf resin explaining a relatively high degree of variation (8.2%, adonis, F = 25.8, P = 0.001). However, those results could be affected by non‐homogeneous dispersion of the data. In fact, specimens without leaf resin are significantly more dispersed (betadisper, P = 0.018), indicating an increased chemical diversity compared to plants with resinous leaves, which are more specialized (Table S7). Notably, no strong associations between leaf chemistry and leaf hairiness nor pollination syndrome are evident (explained variances of 1.4% [adonis, F = 4.16, P = 0.001] and 1.1% [adonis, F = 3.32, P = 0.002], respectively). Univariate testing based on a generalized linear model and subsequent indicator species analysis revealed five chemical families that are significantly associated with bird pollination (Table S8). Those families included a large group of flavonoids (chemical family 9) that harbors the identified 7‐O‐methylated flavonoid cirsimaritin (compound KU036‐6‐1). Future work is needed to explore the presence of these compounds in other plant organs (i.e., flowers, fruits) and a possible correlation to pollination strategy (i.e., guiding birds through flower color) (Panche et al., 2016).
The majority of species in Myoporeae (i.e., phylogenetic clade H) are, with few exceptions including Bontia, Myoporum, and a small number of Eremophila species, distributed in arid/semi‐arid regions of Australia (Figure S9, for a visual representation of a number of major climatic variables in Australia see Figure S10). The complex evolutionary history and spatially heterogenous nature of Australia’s arid zone is now being recognized (Byrne et al., 2008; Murphy et al., 2019), although dividing the arid biome into smaller areas for biogeographic analysis remains problematic (Ebach and Murphy, 2020). In the present study, clear geographic patterns emerge between phylogenetic clades A–H (Figure S9). While phylogenetic clade H is clearly widespread and mainly arid in distribution, the early diverging lineages of the phylogeny (clades A–G) have more coastal/mesic distributions. Within phylogenetic clade H, differences in species distributions between subclades are observed (Figure S11) that can be attributed to the different patterns between the dominant HMC of the heatmap analysis. For example, species in phylogenetic clades H2, H4, H6, H7, H11, and H12 are all distributed in the central arid region of Australia, compared to other subclades that occur in more peripheral Southern or Eastern arid regions. The central arid phylogenetic clades share the unique HMC IX and X chemistry associated with leaf resin, which is largely absent from other lineages. One exception to this is phylogenetic clade H14, which does show some HMC X chemistry and is also mostly resinous but occurs in the Southwest arid region of Western Australia. Unlike all central arid‐distributed phylogenetic subclades, H14 also exhibits unique HMC V chemistry not seen anywhere else in the tribe. The exceptional association of HMC V chemistry with clade H14 is considered in greater detail below.
From the observed chemo‐evolutionary relationships, we envision a chemistry‐based adaptive evolution of species, particularly from phylogenetic clades H2, H4, H6, H7, H11, and H12, associated with the presence of leaf resin. Further molecular work is needed to resolve relationships between these clades, but they may constitute a single evolutionary lineage adapted for the central/Western arid region of Australia. The large number of closely related species in phylogenetic clades H11 and H12 in particular, paired with short branch lengths and a lack of resolution between species relative to other lineages of the phylogeny, suggests that recent and rapid speciation may be responsible for the species diversity. The association between these species and the unique HMC IX‐ and X‐related serrulatane‐ and viscidane‐type diterpenoids, possibly all localized within the leaf resins (Gericke et al., 2020), may have afforded an essential adaptive advantage in the arms race against arid zone herbivores and pathogens, augmenting the number of specialized diterpenoids formed (Becerra, 2007; Richards et al., 2015). A recent chemo‐evolutionary adaptation study in the cosmopolitan Euphorbia genus advocated that greater herbivory pressure resulted in a highly diversified content of toxic diterpenoids in the Afro‐Eurasian geographic region, where specialized herbivores co‐occur, in contrast to the Americas, where specialized herbivores are absent (Ernst et al., 2019b). Evidence suggests that the Australian arid biome is relatively recent in origin and developed within the past 5–10 million years (Byrne et al., 2008; Crisp et al., 2004). The contrasting geographic species distributions between the largely non‐arid phylogenetic clades A–G at the base of the phylogeny and the mainly arid distribution of phylogenetic clade H (Figure S11) support a hypothesis of a relatively recent diversification of Myoporeae in the arid biome coinciding with the evolution of the unique diterpenoid chemistry represented in HMC IX, X, and V. Following diversification into the arid biome, evolution of a resinous protective layer on the leaf surfaces may offer the additional adaptive advantage of reducing water loss via evapotranspiration and protecting leaves from UV and thermal damage by increased reflection of the solar irradiation (Langenheim, 2004). Species without leaf resin may utilize spines and hairiness to deter insects as an alternative to the presence of terpenoids. In addition, the pores in the cell wall may also be fewer or smaller to limit water loss. Species in phylogenetic clades H3 and H10 lack leaf resin and the associated terpene chemistry discussed above. However, these species are characterized with the predominant presence of leaf hairiness and with an additional set of chemistry present in HMC III and IV, respectively. Both HMCs comprise chemical families related to terpenoids as well, in particular monoterpenoid derivatives such as the identified reference compound iridoid glucoside and globularin found in chemical family 3 of HMC III, which are known insect deterrents (Dobler et al., 2011).
Chemo‐evolutionary relationships of serrulatane‐ and viscidane‐type diterpenoids across Myoporeae
We found prenol lipids as the dominant chemical identity within the Myoporeae molecular network, representing 56% of the nodes (1913 nodes) within the 100 largest chemical families. Indeed, the importance of terpenoid‐related metabolism in Myoporeae is suggested by the prevalence and patterns of occurrence of chemical families containing serrulatane‐ and viscidane‐type diterpenoids in several clades (Figure 4). To further investigate this highly diverse family of diterpenoids (Singab et al., 2013), we focused on all chemical families within the connected molecular network associated with either serrulatane or viscidane chemistry. Our analysis led to dereplication of 30 distinct chemical families with these diterpenoid scaffolds, involving 1444 nodes (29% of the connected molecular network). Notably, no overlap of these diterpenoid classes within a chemical family was found, despite the structural similarity of the core structures based on a prenyl tail attached to a bicyclic head. Heatmap (Figure S12) and tanglegram analyses (Figure S3b) of this specific chemical space revealed a bloom of metabolite diversity among phylogenetic clades H2, H4, H6, H7, and H11–H14. This stands in contrast to the less diversified chemistry found in other species of clade H and in clades A–G. This result supports the observed division of species into TMC A and B and emphasizes their role as important factors in understanding chemo‐evolutionary relationships within Myoporeae. The analysis corroborates the co‐occurrence of serrulatane and viscidane diterpenoids with the presence of leaf resin, suggesting important physiological roles of the presence of these compounds at the leaf surface. This finding is supported by our recent study, which investigated the underlying biosynthetic pathways in Eremophila lucida, Eremophila drummondii, and Eremophila denticulata subsp. trisulcata (Gericke et al., 2020), showing that serrulatane and viscidane diterpenoids are concentrated at the resinous leaf surface and synthesized within specialized glandular trichomes embedded within the epidermis. The physiological roles of these diterpenoids are not yet known but could potentially be involved in pathogen defense due to their antibacterial properties (Veneziani et al., 2017) or UV protection based on either absorption‐ or resin‐mediated increased leaf reflectance (Dudek et al., 2016).
Phylogenetic clade H14 presents a unique chemical signature, which specifically comprises HMC V (Figure 4). Two of the dominant chemical families found in HMC V (10 and 22) were dereplicated as serrulatane diterpenoid‐related. Both chemical families show a similar network topology, with a few nodes being found in several species of varying phylogenetic background, besides multiple nodes solely represented by phylogenetic clade H14 (Figure 5). Substructural motif blooms were found within these two chemical families, indicating chemical diversification of 7,8,16‐trihydroxyserrulat‐19‐oic acid (compound KU006‐14) and 8‐hydroxyserrulat‐14‐en‐19‐oic acid (compound KU036‐12) of chemical families 10 and 22, respectively. Besides some reliable high‐level dereplication events, the wide range of metabolite masses and the presence of Eremophila‐specific substructural motifs indicate a large unknown chemical space of serrulatane chemistry, which has yet to be annotated. Based on the chemical information about serrulatane diterpenoids determined within these two chemical families, we attempted to annotate unknown substructural motifs using the combined information from the biochemical reaction prediction tool MetWork (Beauxis and Genta‐Jouve) and the in silico MS2 spectrum prediction tool CFM‐ID (Djoumbou‐Feunang et al., 2019). With this approach we were able to suggest a substructure for the ‘Eremophila motif 440’ that is predominantly present in chemical family 10 and likely relates to the double hydroxylated carboxylic acid of the bicyclic ring present in serrulatane diterpenoids (Figure S13a). This suggested substructural motif overlaps with the MS2LDA spectra and predicted biochemical reactions based on annotated serrulatane diterpenoid structures present in this chemical family. Hereby we found that motif 440 is lost upon dehydroxylation at the aromatic ring, underlying these specific modifications as the inherent characteristics of this motif (Figure S13b). The insufficient dereplication within chemical families 10 and 22 did not allow us to annotate further substructural motifs; however, future chemical annotation of metabolites in Eremophila and related species shall help revealing the nature of these motifs, which display diverse spectral information with gain and loss of small and large fragments (Figure S14). Most of the species within phylogenetic clade H14 have resinous leaves, as do other species from clades H11 and H12 that also correspond to the diterpenoid‐enriched TMC B. The majority of species placed in clade H14 are part of TMC B2, with seven species falling in various parts of TMC A as the exceptions. Species within TMC B2 have resinous leaves, while six of the seven species placed in TMC A have non‐resinous leaves. The sole exception to this is Eremophila subfloccosa subsp. subfloccosa, which has resinous leaves and is found in TMC A3. When specifically focusing the metabolic cluster analysis on all chemical families within the connected molecular network associated with serrulatane and viscidane chemistry, E. subfloccosa subsp. subfloccosa does in fact localize within the resinous members of phylogenetic clade H14 (Figure S3b). Furthermore, none of the non‐resinous species found in clade H14 share features from the serrulatane‐related chemical families in HMC V (10 and 22), underlining the relationship between serrulatane formation and accumulation of leaf resins.
Figure 5.
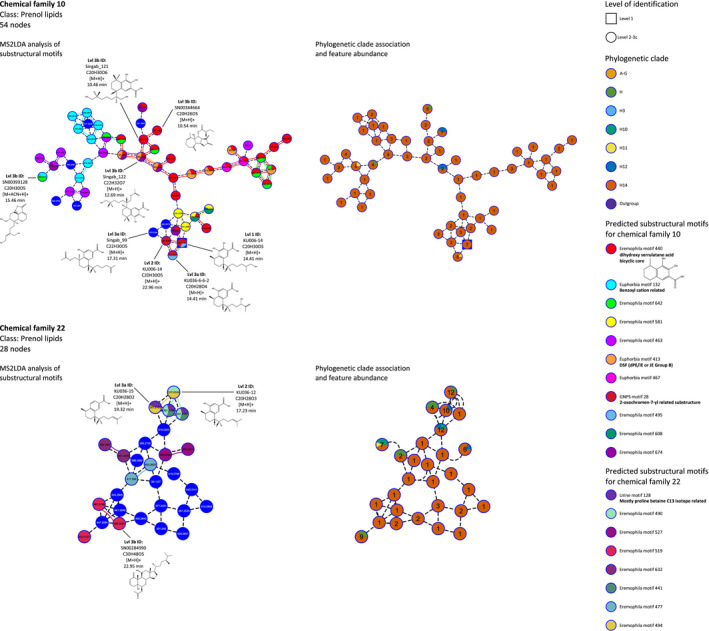
In‐depth analysis of selected serrulatane diterpenoid‐related chemical families.
Detailed analysis of serrulatane diterpenoid‐related chemical families 10 and 22. On the left, chemical families are displayed with detected metabolite mass (Da) information for each node as well as individual dereplication events, which are highlighted with selected spectrometric and structural information. In addition, MS2LDA‐based substructure analysis is shown by color coded pie charts on each node, which point out areas of shared spectral motifs. On the right side, investigated chemical families harboring information about phylogenetic clade associations are shown, as displayed for every node with colored pie charts. Also the total amount of occurrences of each putative metabolite in the dataset is presented for every node.
Phylogenetic clade H14 stands out as an interesting group within tribe Myoporeae because it harbors morphologically highly diverse Eremophila species, a unique chemical signature, and strongly associated antibacterial properties. Notably, there are no published instances of traditional medicinal use for any species of Eremophila placed in clade H14. Both phylogenetic and metabolic cluster analyses support the unexpected placement of morphologically diverse species from three different sections of Eremophila (sensu (Chinnock, 2007)) in phylogenetic clade H14. Two of these sections (Stenochilus and Virides) comprise morphologically similar species, while the third section (Australophilae) contains species that vary considerably from the other two. Morphologically, species in clade H14 have not previously been considered as closely related; however, strong phylogenetic support for this lineage, paired with the unique metabolome identified in the current study, has allowed these relationships to be revealed. This case underlines the utility of combining molecular phylogenetics and metabolite profiling: the observed distribution of particular chemical families supported unexpected phylogenetic relationships and furthermore raises questions about the potential interplay between specialized metabolism and species diversification for future investigation (Afzan et al., 2019).
The molecular networking approach facilitates discovery of novel specialized metabolites
Our approach guides drug isolation efforts by narrowing the large number of metabolites from a plant tribe down to chemical families and predicting chemical classification. This information can be used to target unknown chemical analogues within chemical families with known bioactivities of interest. In addition, functional annotation with the results of antimicrobial assays of crude plant extracts can guide the selection process to uncover novel chemical families not previously considered. A univariate test based on a generalized linear model including tested specimens revealed chemical families significantly associated with antibacterial activity that are solely from the terpenoid‐enriched HMC V and X. A subsequent indicator species analysis further associated these families with positive activity, highlighting them as interesting targets for antibacterial drug discovery (Table S8). When focusing on bioactive serrulatane‐ and viscidane‐type diterpenoids, many of the chemical families are not well structurally dereplicated and are thus excellent targets for drug discovery. In fact, 20 out of 30 chemical families that are part of this particular chemical space contain no level 1 identified metabolite, providing a source of unknown serrulatane or viscidane analogues (Figure S12). To maximize investigated chemical diversity while minimizing drug isolation efforts, we can use the heatmap analysis to identify ideal species for sampling. Using this approach, five plant species (Eremophila galeata, Eremophila enata, Eremophila spectabilis subsp. brevis, Eremophila margarethae, and Eremophila glabra subsp. Paynes Find) out of a total 206 species were shown to cover up to 18% of the observed chemical space of viscidane‐ and serrulatane‐type diterpenoids, accounting for 1444 respective features in total. Other well‐dereplicated chemical families have been chemically classified and should be explored to further enrich our knowledge of the highly diverse diterpenoid structures present. For example, chemical family 10 was found to contain the highly oxygenated serrulatane compound KU006‐14 besides other identified analogues (Figure 5). KU006‐14 was only found in seven of the 23 total species in phylogenetic clade H14, and thus represents a specific branch of the serrulatane‐related chemical space. Compound KU006‐14 is identical to 7,8,16‐trihydroxyserrulat‐19‐oic acid, which we previously isolated from E. drummondii, and carries a hydroxylation at C‐16 as a desired target for substitutions to obtain derivatives with increased Type 2 antidiabetic activity (Wubshet et al., 2016). Thus the C‐16 positioned 3‐methylbutanoyl ester drastically decreased the IC50 of PTP1B inhibition from 1260 ± 560 µm (KU006‐14) to 3.44 ± 0.88 µm (Wubshet et al., 2016). A feature with the corresponding mass ([M+H]+ of 435.2739 Da) to this chemical analogue can be found in chemical family 10 in close proximity to compound KU006‐14. Further exploration of chemical family 10‐associated diterpenoids and subsequent testing for their antidiabetic activities may reveal the presence of even better drug candidates in other species of phylogenetic clade H14. Additional synthetic functionalization may further extend the potential biological activity of compound KU006‐14‐related metabolites. This has been documented for the close analogue 3,7,8‐trihydroxyserrulat‐14‐en‐19‐oic acid (Kumar et al., 2017). This compound did not show any antimalarial activity, whereas a number of derived amides did.
Our results show how our molecular networking pipeline can aid in the identification of both plant species and chemical families with potential as sources of new bioactive molecules. Chemical families with low levels of metabolite identification based on in silico spectral support are easily determined and represent a prime source to unfold the rich chemical ‘dark matter’ in Myoporeae (da Silva et al., 2015). A single example is chemical family 5, which includes 110 nodes and was dereplicated as fatty acid‐related. Notably, this prediction is solely based on level 3a identifications, and therefore warrants research to identify the structures of some of the features in the molecular network. The median mass of 332.2732 Da and the retention time of 17.8 min of this chemical family are comparable with known diterpenoids in this study and further suggest an interesting source for potential drug candidates. Additionally, this chemistry is mainly localized to species of phylogenetic clades A–G, indicating its role in specialized metabolism. Notably, the resinous Glycocystis beckeri and Myoporum bateae contribute significantly to the chemical space in chemical family 5 and thus represent suitable targets for investigation. Additionally, these two species also harbor flavone‐related features from chemical family 11, which accumulate mostly in clades A–G.
Australian Aboriginal Peoples use herbal extracts from some Eremophila species as integrated components of their traditional medicine (Chinnock, 2007; Richmond, 1993; Richmond and Ghisalberti, 1994). These herbal extracts contain a multiplicity of bioactive natural products that individually or in combination could provide the perceived curing effects. In an attempt to begin to associate the presence of specific bioactive natural products or combinations thereof to the beneficial effects, species with documented Aboriginal uses were highlighted in our current study. To underline this approach, measured antibacterial effects of herbal extracts against human pathogenic bacterial species and of individual isolated natural products were also integrated into this investigation (Table S1). Traditional medicinal uses of herbal extracts were widespread throughout Myoporeae. Interestingly, no use of Eremophila species from phylogenetic clade H14 was found in the literature. This contrasts with the number of reports documenting a high abundance of herbal extracts and isolated diterpenoids possessing antibacterial activity within this subclade (Lyddiard and Greatrex, 2018; Ndi et al., 2007b). This could indicate that the diversity of serrulatanes in clade H14 somehow is associated with negative effects in traditional medicine. However, the use of plants within Aboriginal traditional medicine is likely to be underreported in the literature. It is recognized that much traditional knowledge was lost with the break‐up of traditional societies and the denial of access to land and natural resources for Aboriginal Peoples following the British colonization of Australia (Smith, 1991; Zola and Gott, 1992). Furthermore, some traditional custodians may have chosen not to have this information recorded in the public domain. Therefore, the absence of recorded traditional use of Eremophila species in clade H14 does not mean that some of these species were not used by a specific group at some point in time. Nevertheless, from the records it is clear that E. alternifolia had and continues to have a prominent medicinal role (Richmond, 1993; Singab et al., 2013; Smith, 1991), and it is recognized in our current study for its rich flavone‐related chemistry. A second prominent species is Eremophila cuneifolia (Richmond, 1993; Singab et al., 2013) with a distinct elevated compound diversity in terpenoid‐related chemical families 23 and 95, which are mostly absent in other species. Thus, successful functional annotations to a specific molecular network could enable assignment of metabolites present to specific biological activities, morphological traits, and geographical distributions and enable targeted drug discovery approaches (Fox Ramos et al., 2019; Nothias et al., 2018).
CONCLUSION
The present study highlights the power of combining large‐scale molecular networking and phylogenetic analyses to investigate chemo‐evolutionary diversification that involves specialized diterpenoid chemistry associated with resin development in Myoporeae. By integrating functional annotations in our analysis, we hypothesize that this specialized chemistry may complement the transpiration barrier function of the resin with pathogen and herbivore defense as well as UV protection. To shed further light on the broader area of arid zone ecosystem evolution, ecological and phylogenetic information for associated insects could be integrated into the chemo‐evolutionary framework developed here for Myoporeae. A number of studies have shown several insect groups that occur almost exclusively with Eremophila (Cassis and Symonds, 2008; Symonds and Cassis, 2014). Further research in this area may help us to understand the links between chemistry and species radiation (both plants and insects) as well as regional and fine‐scale patterns of diversification.
Despite great progress in illuminating the chemical space of Myoporeae, there are still chemical families of unknown character waiting to be elucidated. Our study shows the state‐of‐the‐art computational tools that can be used to target drug discovery approaches by giving an idea about the identity of the observed chemical space. Furthermore, this information can be correlated with transcriptomics, genomics, or proteomics data to elucidate the underlying biosynthetic machinery for chemical families of interest (Fox Ramos et al., 2019). Additionally, the chemical component of the present study focused solely on leaf chemistry. However, ongoing research in Eremophila and other plant groups has revealed that other plant organs may also be rich sources of distinct bioactive metabolites. An example is the antimalarial microthecaline A, a novel quinoline–serrulatane alkaloid, which was found in roots of Eremophila microtheca and Myoporum insulare (Kjaerulff et al., 2020; Kumar et al., 2018).
By establishing a chemo‐evolutionary framework and enriching it with functional annotations, we present a systematic outline of chemistry and evolution in Myoporeae and lay foundations for further interdisciplinary research. We encourage the use and further extension of this dataset through functional annotations to explore chemo‐evolutionary relationships and to select compelling chemical families for natural product isolation to describe the unknown aspects of Myoporeae chemistry. Furthermore, we provide the exhaustive library of 76 reference compound spectra isolated from Myoporeae species to the natural products community to help annotating this interesting chemistry in other metabolomics studies.
Eremophilas are culturally important plants for many of Australia's First Peoples, the Aboriginal peoples. If you use the information here provided to make commercial products, we urge you to strongly consider benefit sharing with the Aboriginal communities or groups in the areas where these species grow. We acknowledge that this work took place on the lands of Aboriginal peoples who are the custodians of this land, and acknowledge and pay our respects to their Elders past and present.
EXPERIMENTAL PROCEDURES
Collection of plant material
Leaf tissue and herbarium voucher specimens were collected from field and cultivated collections from across Australia. A cultivated specimen of Bontia daphnoides was sent from Fairchild Tropical Botanic Garden, Florida. Collection details, herbarium voucher numbers, and GenBank accession numbers are provided in Table S1. Leaf material was picked fresh and stored immediately in silica beads. The same silica dried leaf material was used for DNA and phytochemical extractions.
Phylogenetic analysis
Total genomic DNA was isolated using a modified cetyltrimethylammonium bromide protocol (Shepherd and McLay, 2011). Library preparation was completed ‘in‐house’, using a version of the sample preparation protocol outlined in Schuster et al. (2018). Samples were sequenced using both Illumina HiSeq2000 (2 × 125 bp) and Illumina NextSeq500 (2 × 150 bp) sequencing platforms, based at AgriBio, Centre for AgriBioscience (La Trobe University) and the Walter and Eliza Hall Institute (WEHI) in Melbourne, Australia. Raw read data of total genomic DNA were de novo assembled using CLC Genomics Workbench version 8.5.1 (https://www.qiagenbioinformatics.com/) with default settings. Resulting contigs were imported into Geneious version 9.1.8 (http://www.geneious.com) (Kearse et al., 2012), where the nuclear ribosomal cistron was constructed for each sample, initially using the partial internal transcribed spacer (ITS)/external transcribed spacer (ETS) sequence available for Eremophila macdonnellii on GenBank (DQ444239) as a reference. Contigs were mapped to E. macdonnellii using custom sensitivity settings; gaps allowed were set to a maximum of 20% per read, the maximum gap size was 3000, and 2–3 iterations were used. Total raw reads were then mapped back to the consensus sequence and a final consensus sequence extracted using a consensus threshold of 75% to confirm sequence accuracy. Nuclear ribosomal cistron length varied between samples from 5976 to 7863 base pairs. Differences in sequence lengths were largely due to variation in DNA read coverage of the ETS/non‐transcribed spacer (NTS) regions. All samples contained complete coverage of 18S, ITS1, 5.8S, ITS2, and 26S nuclear ribosomal DNA regions. Sequences were aligned in Geneious using the MAFFT pairwise alignment plug‐in (mafft version 7.222; Katoh and Standley, 2013) with default settings. The alignment was assessed by eye and any adjustments were made manually. After exclusion of poorly aligned sequence ends, the 291 nuclear ribosomal sequences resulted in an aligned matrix of 6568 bp. This alignment is available in TreeBase (study accession number 26197). Phylogenetic analysis was completed using MrBayes version 2.3 (Ronquist et al., 2012). The alignment was partitioned into six character sets representing coding ribosomal DNA sequences (18S, 5.8S, and 28S genes) and intergenic spacers (ITS1, ITS2, and ETS+NTS). Models of evolution for each partition were estimated using the Bayesian information criterion in IQ‐TREE version 1.6.12 (Nguyen et al., 2015). Where exact models were not available in mrbayes equivalent models were selected (18S: GTR+I+G, 5.8S: K2P+I, 26S: GTR+I+G, ITS1: SYM+G, ITS2: SYM+I+G, NTS: GTR+I+G). The Bayesian inference analysis was run for 15 million generations with unlinked partitions and a tree sampling frequency of 1000 to estimate posterior probabilities. The average standard deviation of split frequencies reached a value less than 0.01 during this analysis, with convergence of Markov chain Monte Carlo chains checked in Tracer version 1.6 (Rambaut et al., 2018). First, 25% of the trees were discarded as burn‐in, then a consensus tree was generated, and Bayesian posterior probabilities were estimated for nodes from the consensus tree (Figure S15).
Metabolite analysis
Ground and dried leaf tissues were extracted with 50% acetonitrile (supplemented with 2 ppm forskolin), while shaking incubation at 25°C for 2 h. Acetonitrile extracts were filtered using a 0.22‐µm 96‐well filter plate (Merck Millipore, Darmstadt, Germany) and analyzed using an Ultimate 3000 UHPLC+ Focused system (Dionex Corporation, Sunnyvale, CA, USA) coupled to a Bruker Compact ESI‐QTOF‐MS (Bruker, Billerica, MA, USA) system. Samples were separated on a Kinetex XB‐C18 column (100 × 2.1 mm internal diameter, 1.7 µm particle size, 100 Å pore size; Phenomenex Inc., Torrance, CA, USA) maintained at 40°C with a flow rate of 0.3 ml min−1 and a mobile phase consisting of 0.05% (v/v) formic acid in water (solvent A) and 0.05% (v/v) formic acid in acetonitrile (solvent B). The LC method was as follows: 0–1 min, 10% B; 1–23 min, linear increase from 10 to 100% B; 23–25 min, 100% B; 25–25.5 min, 100–20%; 25.5–30.5 min, linear decrease to 10% B. Mass spectra were acquired in positive ion mode over a scan range of m/z 50–1200 with the following ESI and MS settings: capillary voltage, 4000 V; end plate offset, 500 V; dry gas temperature, 220°C; dry gas flow, 8 L min−1; nebulizer pressure, 2 bar; in‐source collision‐induced dissociation (CID) energy, 0 eV; hexapole radiofrequency, 50 Vpp; quadrupole ion energy, 4 eV; collision cell energy, 7 eV. Samples were further subjected to untargeted LC‐MS/MS using a collision cell energy of 27 eV. Quality control samples were prepared from a pool of different extract samples and run after a sequence of 22 samples as well as before and after the total run. Raw chromatogram data were calibrated using an internal sodium formate standard and subsequently exported in .mzML format using DataAnalysis 4.1 (Bruker). Further processing of the raw chromatogram data was conducted using MZmine2 (v.2.5.3) (Pluskal et al., 2010). At first, a signal intensity noise cutoff of 1000 and 100 was applied to MS1 and MS2 data, respectively. In addition, only scans between 0.5 and 24 min retention time were considered. Chromatograms have been built using the ‘ADAP chromatogram builder’ with an m/z tolerance of 0.003 Da (5 ppm) and a signal intensity threshold of 20 000. The generated extracted ion chromatograms were deconvoluted into individual peaks using the ‘local minimum search’ module. After isotopes were removed, the feature list was aligned using the ‘joint aligner’ tool using an m/z tolerance of 0.006 Da (10 ppm) and a retention time tolerance of 0.1 min. Subsequently, individual metabolites were identified by comparing them to MS2 data of an in‐house spectral database (level 1 identification includes m/z, retention time, and MS2 similarity). This library includes 76 reference compounds commercially sourced or isolated from different Eremophila species during prior studies (Pedersen et al., 2020; Zhao et al., 2019; Zhao et al., unpublished data) (Table S3), which were analyzed with the same MS system as described above. Missing data points were added using the ‘gap filler’ tool and afterwards intensities below 1000 were set to 0 by applying a peak filter. The resulting peak list was further subjected to a quality control that was conducted by a principal component analysis comprising all species and quality control samples on top of the gap‐filled (but not peak‐filtered) peak list. In this projection, all quality control samples are found in close proximity, indicating reliable data acquisition over time (Figure S16). Finally, background features that were shared between the samples and quality controls were removed manually, which includes the internal standard forskolin as well (data kept for normalization). The generated peak list was exported twice, as a .csv file (containing peak signal intensity) for further processing in R script and via the ‘GNPS‐FBMN export’ module for subsequent upload to the GNPS server. A detailed description of the MZmine2 processing parameters used can be found in Table S9. A molecular network was created with the Feature‐Based Molecular Networking workflow (Nothias et al., 2020) on GNPS (https://gnps.ucsd.edu) (Wang et al., 2016). The data were filtered by removing all MS2 fragment ions within ±17 Da of the precursor m/z. MS2 spectra were window‐filtered by choosing only the top six fragment ions in the ±50‐Da window throughout the spectrum. The precursor ion mass tolerance was set to 0.02 Da and the MS2 fragment ion tolerance to 0.02 Da. A molecular network was then built where edges were filtered to have a cosine score above 0.8 and more than eight matched peaks. Furthermore, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top five most similar nodes. Finally, the maximum size of a chemical family was set to 0 (no size limitation) and the lowest scoring edges were removed from chemical families until the chemical family size was below this threshold. The spectra in the network were then searched against GNPS spectral libraries (Horai et al., 2010; Wang et al., 2016) as well as an in‐house library including 76 reference spectra (level 2 identification includes MS2 similarity). The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least six matched peaks. The molecular networks were visualized using Cytoscape software (v3.7.2) (Shannon, 2003). To predict a consensus of chemical classification for individual chemical families, the NAP tool was applied to the generated molecular network (Da Silva et al., 2018). Thereby, multiple public in silico spectral databases were used, i.e., GNPS, SUPNAT, NPAtlas, CHEBI, and DRUGBANK. Additionally, an Eremophila‐specific in‐house in silico fragmentation database was used that contains 293 metabolite structures that have been characterized in Eremophila species during prior studies (Pedersen et al., 2019, 2020; Singab et al., 2013; Zhao et al., 2019; Zhao et al., unpublished data). The in silico fragmentation‐based dereplication results were categorized by their reliability, with SMILES from Fusion, Consensus, and MetFrag algorithms corresponding to level 3a, 3b, and 3c identification, respectively. Additionally, insight into substructural information was obtained using the MS2LDA motif search via the GNPS server. MS2LDA allows annotation of smaller substructures shared by metabolites of a chemical family within a molecular network (van der Hooft et al., 2016). Interpretation of novel ‘Eremophila motifs’ in chemical families 10 and 22 was based on the combined information of biochemical and MS2 spectrum predictions using the tools MetWork (Beauxis and Genta‐Jouve) and CFM‐ID 3.0 (Djoumbou‐Feunang et al., 2019), respectively. For this, the spectral data for corresponding features were extracted from the GNPS analysis and uploaded to the MetWork server as a .mgf file and subsequently analyzed using default settings. Fragment spectrum predictions on annotated serrulatane diterpenoid structures were made using the CFM‐ID webserver under the following settings: Spectra Type ‘ESI’, Ion Mode ‘Positive’, and Adduct Type ‘[M+H]+’. Eventually, the outputs from the different molecular network‐based analyses (FBMN, NAP, and MS2LDA) were joined via the MolNetEnhancer (v15) workflow, which links the inferred structural information to increase the resulting chemical insight obtained from a spectral dataset (Ernst et al., 2019a). A detailed description of the molecular networking pipeline presented in this study can be found in Table S10. The MZmine2 pre‐processed chromatogram data were further processed for downstream analysis using R script (v3.6.1). Thereby, the data were normalized to the median of the internal standard and sample weight. Subsequently, a signal intensity cutoff of 10 000 was applied and resulting features that had no occurrence anymorewere removed, resulting in a normalized dataset of 10 696 features, removing 507 features in total that are mostly present as singletons within the molecular network (70%). Eventually, every peak signal intensity above 0 was assigned the value ‘1’ to create a binary matrix. To add the information gained from the molecular network, the 100 largest chemical families with at least nine nodes (Table S11) were isolated and combined with the processed chromatogram data to generate a new data matrix. In this way, a ‘continuous’ dataset that contains the number of features that are shared of a particular chemical family for each species was generated. Additionally, a ‘categorical’ dataset was generated that contains solely the value ‘1’ for a species that shares at least one node of a chemical family and ‘0’ for not having a single node (Figure 4). Based on the categorical dataset, a distance matrix was generated using Jaccard distance, which was subjected to a hierarchical cluster analysis using the ‘ward.d2’ agglomeration method. Using the ‘ape’ package (v5.3), this analysis was converted to a dendrogram, which was then compared to the dendrogram derived from the molecular phylogeny via a tanglegram analysis (‘dendextend’ package v1.12.0) (Galili, 2015). The resulting tanglegram was untangled using the ‘step2side’ algorithm, which utilizes a greedy stepwise rotation of the two dendrograms to find the best match, while the resulting rearranged molecular phylogeny was used for further heatmap analyses. Using the R package ‘ComplexHeatmap’, a heatmap was generated based on the ‘categorical’ dataset using Jaccard distance and the ‘ward.d2’ agglomeration method (km = 10 000), which clusters the presence/absence of chemical families according to phylogenetic similarity, while the order of specimens in the phylogeny is fixed. A ‘continuous’ heatmap analysis is based on the cluster generated by the ‘categorical’ analysis and highlights the total number of putative metabolites from a chemical family for each specimen (Figure S4). Combined detailed information of tanglegram and heatmap analyses is found in Table S12. Additional information about chemical classification (Tables S13 and S14) and leaf morphology (resin and hairiness), environmental factors (pollination and geographical distribution), and medicinal properties (traditional medicinal uses and antibacterial studies) were aligned to the heatmap analyses. Another heatmap analysis focused solely on serrulatane and viscidane diterpenoid‐related chemical families. In that regard, all chemical families (down to two nodes) that contain at least one corresponding dereplication hit of level 1–3c were selected manually from the global molecular network (Table S15, Table S16). These 30 chemical families were subjected to a tanglegram analysis as described above (Figure S3b) as well as a heatmap analysis that clustered the ‘continuous’ chemical information using Euclidean distance and the ‘complete’ agglomeration method (Figure S12). Another tanglegram was conducted based on the presence or absence of all 10 696 individual metabolites after normalization (Figure S3a). To address the chemical identity of selected singleton features, we used the machine learning‐based in silico structure prediction software sirius with the following adjustments besides default settings: sirius – H, C, and O elements plus [M+H] and [M+Na] ionization; CSI:FingerID – Bio Database and [M+H], [M+Na], and [M‐H20+H] ionization; CANOPUS – default settings (Duhrkop et al., 2015; Dührkop et al., 2019, 2021).
Statistical analysis
To test for overall significant differences of functional annotations among the investigated specimens, we used the non‐parametric permutational multivariate statistical test permanova (Anderson, 2001). This method uses a dissimilarity metric to calculate differences between annotation object classes. For permanova, the null hypothesis is that the metric centroid does not differ between groups. Using R script, permanova was performed with the adonis2() function from the vegan package (Oksanen et al., 2019). This analysis was based on the above described categorical dataset, for which the dissimilarity metric was generated by the vegdist() function using Jaccard distance. permanova is sensitive to differences in data dispersion within groups and may therefore confuse within‐group variation with among‐group variation (Anderson and Walsh, 2013). To test if groups differed in their dispersion, we used the betadisper() function with the null hypothesis that the average within‐group dispersion is the same in all groups. In each test, the number of permutations was set to 999. For all analyses, we considered a P‐value threshold of ≤0.05 significant. We conducted these tests on the full dataset (100 chemical families) to test the significance of individual functional annotations such as leaf resin, leaf hairiness, antibacterial activity, pollination, biogeography, and general clades (‘O’: outgroup, ‘AG’: clades A to G, ‘H3’: clade H3, ‘H10’: clade H10, ‘H11’: clade H11, ‘H12’: clade H12, ‘H14’: clade H14, and ‘H’: remaining clade H‐associated taxa), whereby specimens with missing values were excluded. Since records for Aboriginal usage are scarce, we excluded it from all statistical analyses. We used generalized linear models to test for significant differences of functional annotations among the investigated specimens within an HMC background (Warton et al., 2012). Individual models were generated for each HMC–annotation combination using the clogloc model, while excluding specimens with missing functional annotation. Each test was conducted using the anova.manyglm() function from the mvabund package (Wang et al., 2020). In each test, the number of bootstrap iterations was set to 999 and we used the Monte Carlo resampling method. For all analyses, we considered a P‐value threshold of ≤0.05 significant. To assess individual chemical families as overall significant drivers for a functional annotation, a subsequent univariate test was conducted for the generated generalized linear models to receive individual P‐values. An additional indicator species analysis using the indval() function (labdsv package; Roberts, 2019) was applied to indicate the direction of annotation (level) for each chemical family (Table S8).
Visualizing species geographic distributions
Geographic distribution maps representing species present in tanglegram metabolite clusters or phylogenetic clades were generated using species occurrence records downloaded from the Australasian Virtual Herbarium (https://avh.ala.org.au). All species occurrence records used in mapping figures are available from DOI links provided below. Species distributions were overlayed on the Crisp and Cook (2013) published map of Australian biomes (Crisp and Cook, 2013), and any species spanning two or more of these biomes was assessed as widespread for mapping purposes. To allow for the clear visualization of biogeographic results and mitigate the effects of widespread taxa swamping the biogeographic patterns, widespread species (E. alternifolia, Eremophila bignoniiflora, E. deserti, Eremophila latrobei subsp. glabra, Eremophila longifolia, Eremophila mitchellii, Eremophila maculata subsp. maculata, Eremophila polyclada, Myoporum acuminatum, and Myoporum montanum) were removed from maps (as per Murphy et al., 2019).
AUTHOR CONTRIBUTIONS
OG, RMF, and BLM designed the study. RMF, MJB, and BJB collected, identified, and sampled Eremophila and related species from field and cultivated collections from across Australia. RMF and MJB performed taxonomic description and phylogenetic analyses of these specimens. OG and AMH prepared the plant extracts and performed the LC‐QTOF‐HRMS analysis. OG conducted the mass spectral data processing and quality check as well as subsequent molecular networking analysis. All reference compounds derived from Eremophila species were isolated, structurally characterized, and provided by DS. OG established the in‐house reference compound and in silico spectral databases and performed dereplication of the molecular network including the MS2LDA substructural analysis. OG established the computational pipeline to establish and analyze the chemo‐evolutionary framework in Myoporeae and wrote all scripts used to conduct data processing as well as cluster, tanglegram, heatmap, and statistical analyses. Different authors provided information about functional annotations, leaf resin, hairiness, and pollination (RMF and BJB), antibacterial activity and traditional usage (RMF, SJS, and CPN), and biogeography (RMF and DJM). Species distribution mapping was completed by RMF. OG, RMF, and BLM wrote the manuscript. All authors discussed the results and contributed with comments during the writing process.
CONFLICT OF INTERESTS
The authors declare that there are no competing interests.
Supporting information
Table S1. Specimen details for all samples included in this study. Specimen information includes collection number, species name (authority), Eremophila section, collection location, herbarium voucher number, GenBank accession number, and collected metadata used in this study. Collecting numbers followed by * denote specimens grown in cultivation; where provenance information was known this was recorded in latitude/longitude. Collection initials refer to: RMF (Rachael Fowler), MJB (Michael Bayly), BB (Bevan Buirchell), TMS (Tanja Schuster), FTBG (Fairchild Tropical Botanic Garden), NTBG (National Tropical Botanical Garden). Published records of traditional medicinal use for each species are recorded where available (Barr, 1988; Barr et al., 1993; Bishop Museum, 2020; Bowen, 1975; Clarke, 2013; Clarke, 2014; Clealand and Tindale, 1959; Cunningham et al., 1981; Evans et al., 2010; Isaacs, 1987; Lans, 2006; Lassak and McCarthy, 1983; Latz, 1982; Latz and Green, 1995; Liu et al., 2006; Meggitt, 1962; Ndi et al., 2007b; O’Connell et al., 1983; Richmond, 1993; Richmond and Ghisalberti, 1995; Sadgrove et al., 2016; Sadgrove and Jones, 2013; Silberbauer, 1971; Smith, 1991; Spencer and Gillen, 1969; Tindale, 1937; Tynan, 1979; Webb, 1948; Wong, 1976; Woodworth; Zaleta‐Pinet et al., 2016). Evidence of antimicrobial activity against human bacterial pathogens (Gram‐positive bacteria) for each species is recorded where available (Algreiby et al., 2018; Aminimoghadamfarouj and Nematollahi, 2017; Barnes et al., 2013; Biva et al., 2016; Biva et al., 2019; Evans et al., 2010; Galappathie et al., 2017; Liu et al., 2006; Lyddiard and Greatrex, 2018; Ndi et al., 2007a, 2007b, 2007c; Salama, 2017; Smith et al., 2007). Metadata information is interpreted as follows: leaf resin: ‘0’ – absence, ‘1’ – presence; leaf hairiness: ‘0’ – absence, ‘1’ – presence; pollination: ‘0’ – insect, ‘1’ – bird; traditional use: ‘0’ – no documented use, ‘1’ – use is documented; antibacterial activity: ‘0’ – tested absence of activity, ‘1’ – tested presence of activity; biogeographical distribution: ‘0’ – widespread, ‘1’ – Eremean, ‘2’ – Southwestern temperate, ‘3’ – Southeastern temperate, ‘5’ –non‐Australian; phylogenetic clade association: ‘o’ – outgroup, ‘ag’ – clades A to G, ‘h3’ – clade H3, ‘h10’ – clade H10, ‘h11’ – clade H11, ‘h12’ – clade H12, ‘h14’ – clade H14, and ‘h’ – remaining clade H‐associated taxa.
Table S2. sirius‐based chemical classification of 10 singletons with the highest abundance. The table shows spectral information as well as the results of the chemical annotation of 10 singleton features found to have the highest abundance (peak signal intensity) in the Myoporeae spectral dataset. Chemical annotation was conducted here using sirius software and did not rely on feature‐based networking.
Table S3. List of reference compounds used as in‐house spectral database. Detailed spectrometric and structural information about all reference compounds used in this study.
Table S4. Detailed information for generated tanglegrams in this study. The table shows detailed information about common subbranches found in each tanglegram, which are indicated with a distinct taxon group ID and corresponding specimen ID tag for comparison. Further below are displayed the total number of taxa groups per tanglegram and the corresponding entanglement factor.
Table S5. Statistical analysis of individual heatmap metabolic cluster using general linear models. Generalized linear models were used to test for significant differences of functional annotations among the investigated specimens within a heatmap metabolic cluster (HMC) background. Models were generated for each HMC–attribute combination using the clogloc model, while excluding specimens with missing functional annotation. Each test was conducted using the anova.manyglm() function from the mvabund package. A P‐value of ≤0.05 was considered significant. ‘Res. Df’ – residual degrees of freedom; ‘Df.diff’ – model degrees of freedom; ‘Deviance’ – deviance of a fitted multivariate model object for abundance data.
Table S6. Chemical comparison of tanglegram metabolic cluster (TMC) A and B. Comparison of the chemical content between TMC A and B. The underlying metabolic cluster analysis is based on the 100 largest chemical families of the generated molecular network of Myoporeae. Numbers of specimens from major phylogenetic clades are shown with ‘O’: outgroup, ‘AG’: clades A to G, ‘H3’: clade H3, ‘H10’: clade H10, ‘H11’: clade H11, ‘H12’: clade H12, ‘H14’: clade H14, and ‘H’: remaining clade H‐associated taxa. Dominant chemical families are depicted for each TMC, with ‘CF80’: chemical families present in 80% of the specimens, ‘CF50’ and ‘CF30’: 50% and 30% abundancy of a given family among the given specimens, respectively. Different levels of predicted chemical classification and corresponding scores are shown for each chemical family. If present, level 1 identifications and their associated molecular structures are displayed.
Table S7. Multivariate statistical analysis involving the chemical space of Myoporeae. Permutational multivariate analysis of variance (permanova) was conducted involving all samples associated with a given functional annotation. Results of the Adonis2() and betadisper() functions are shown, which give information about statistical significance and data dispersion of tested attribute levels, respectively.
Table S8. Univariate tests and indicator species analysis of individual chemical families. To determine individual chemical families as overall significantly associated to a functional annotation, a univariate test was conducted using generalized linear models including all specimens with corresponding metadata to receive adjusted P‐values. An additional indicator species analysis using the indval() function was conducted to indicate the direction of annotation (level) for each chemical family.
Table S9. MZmine2 parameters used for raw chromatogram data processing. Detailed overview of parameters used for raw chromatogram data processing done in this study. This was carried out using the software MzMine2 (Pluskal et al., 2010).
Table S10. Molecular networking parameters. Detailed overview of parameters used for the feature‐based molecular networking approach presented in this study. The analysis was carried out using the Global Natural Products Social Molecular Networking (GNPS) toolbox (Wang et al., 2016).
Table S11. List of the 100 largest chemical families derived from the molecular network of Myoporeae. Detailed information about the largest 100 chemical families generated by the molecular networking approach conducted in this study. Chemical family classification is predicted based on the applied dereplication pipeline. Further indicated are total node count and total numbers of different levels of chemical identification determined in this analysis.
Table S12. Phylogenetic and chemical information about the performed metabolic cluster analysis in Myoporeae. Comparison of the chemical content information extracted from determined tanglegram metabolic clusters (TMCs). The underlying metabolic cluster analysis is based on the absence/presence of the 100 largest chemical families of the generated molecular network of Myoporeae. Numbers of specimens from major phylogenetic clades are shown with ‘O’: outgroup, ‘AG’: clades A to G, ‘H3’: clade H3, ‘H10’: clade H10, ‘H11’: clade H11, ‘H12’: clade H12, ‘H14’: clade H14, and ‘H’: remaining clade H‐associated taxa. Dominant chemical families are depicted for each TMC, with ‘CF100’ indicating chemical families present in all species (100%) of the corresponding TMC and ‘CF90’ and ‘CF80’ depicting 90% and 80% abundancy of chemical families among the associated specimens, respectively. Different levels of predicted chemical classification and associated scores are shown. If present, level 1 identifications and their associated molecular structures are displayed.
Table S13. Detailed chemical information about heatmap metabolic clusters. Heatmap metabolic clusters (HMCs) were generated using a categorical heatmap analysis based on the presence/absence of the 100 largest chemical families found in the molecular network of Myoporeae. Information about chemical identity is given based on the conducted dereplication pipeline. Further indicated is spectrometric and structural information about corresponding level 1 identification events.
Table S14. Identified metabolites within the 100 largest chemical families in Myoporeae. Detailed spectrometric and structural information about level 1 and 2 identification events found within the largest 100 chemical families from the molecular network of Myoporeae.
Table S15. Dereplication of serrulatane‐ and viscidane‐related chemical families. A list of 30 chemical families from Myoporeae that have been dereplicated as serrulatane‐ or viscidane‐related. Chemical family classification is predicted based on the applied dereplication pipeline. Further indicated are total node count and total numbers of different levels of chemical identification determined in this analysis as well as mass and retention time median of each chemical family.
Table S16. Predicted serrulatane and viscidane diterpenoid structures in chemical families of Myoporeae. Detailed spectrometric and structural information about serrulatane and viscidane diterpenoid‐related identification events determined within the 30 chemical families within the Myoporeae molecular network that have been found to be associated with this specific chemistry.
Figure S1. Schematic workflow of the chemo‐evolutionary analysis. In total, 291 specimens from species and subspecies of the tribe Myoporeae were investigated regarding their chemical and phylogenetic relationships. LC‐QTOF‐HRMS was applied on crude leaf extracts to generate mass spectrometric data including MS1 and MS2. MZmine2 was used to isolate spectral features from the raw data and align those across all samples to output a feature table that contains the relatively quantified chemical composition of the dataset. Spectral information of those isolated features is then exported and integrated into the feature‐based molecular networking (FBMN) pipeline at GNPS (https://gnps.ucsd.edu). The generated molecular network is composed of nodes and edges that represent individual spectral features and share spectral similarity, respectively. Spectral similar nodes fall into clusters of varying size that are considered as a chemical family of structurally related metabolites. The molecular network provides the foundation for an exhaustive dereplication approach involving public, in‐house, and in silico libraries of reference metabolites for spectral comparison. Dereplicated nodes are structurally annotated and utilized to achieve a prediction of chemical classification for each chemical family by using the Network Annotation Propagation (NAP) method. Using the unsupervised substructure discovery approach MS2LDA, substructural motifs can be predicted within the given spectral data and annotated throughout the molecular network. To compare the chemical profiles of all 291 specimens, the presence (at least one spectral feature present of a corresponding chemical family) and absence (no feature present in the sample) information of chemical families can be used. This approach yields a binary dataset that can be compiled by a metabolic cluster analysis into a dendrogram that displays chemical similarity among the specimens. In contrast, a phylogenetic analysis shows the evolutionary relationships among those specimens and is based on nuclear ribosomal DNA sequencing of leaf tissue from the same specimens. A tanglegram analysis directly compares the analyses by rearranging branches in both dendrograms to reveal similar clustering (i.e., specimens D and E) and thus chemo‐evolutionary relationships. Groups of specimens that display an evolutionary lineage are referred to as phylogenetic clades, while chemical similarity is shared within tanglegram metabolic clusters (TMCs). To extend the view on the chemo‐evolutionary framework, a categorical heatmap analysis is used that clusters the presence/absence of individual chemical families according to their phylogenetic distribution alongside the tanglegram‐refined phylogeny. Heatmap metabolic clusters (HMCs) can be derived from this approach, which represent subsets of chemical families that share a similar phylogenetic signature. Additional functional annotations can be integrated to test metadata for correlation with the chemo‐evolutionary framework that is displayed by the heatmap.
Figure S2. Specimen and peak signal intensity distribution of singleton features. Distribution graphs showing the number of singleton features according to the number of associated specimens and peak signal intensity (detected maximum value for each feature among all specimens). Calculated medians are highlighted by a dotted vertical line and the actual value displayed below each respective graph.
Figure S3. Collection of tanglegram analyses. Conjunction of phylogenetic and metabolic information from 291 species of the tribe Myoporeae visualized by different tanglegram analyses. The nuclear ribosomal DNA‐based phylogeny shows taxa with posterior probability above 95%. Metabolic cluster analyses were conducted based on different datasets: (a) presence/absence of individual spectral features after normalization of the full spectral dataset (10 696 features); (b) presence/absence of the 30 chemical families found to be associated with serrulatane or viscidane diterpenoid metabolism within the generated global molecular network of Myoporeae; (c) peak signal intensity of individual spectral features after normalization of the full spectral dataset (10 696 features); (d) presence/absence of individual spectral features assigned as singletons after normalization; and (e–g) presence/absence of the 100 largest chemical families within the generated molecular network of Myoporeae using an inclusion factor of (e) 2, (f) 3, and (g) 5. Phylogenetic subclades are indicated with the most prevalent ones highlighted by color code. The tanglegram analysis connects the same specimens by a line, which is colored when equal branching of taxa is present in both analyses. Selected species with varying intraspecific chemical variation are highlighted within selected metabolic cluster analyses, with square (E. duttonii), diamond (E. alternifolia), and circle (E. deserti).
Figure S4. Continuous heatmap analysis showing chemo‐evolutionary relationships in Myoporeae. A heatmap analysis was used to put chemical information into an evolutionary context. For that, information about the 100 largest chemical families was compiled, which was derived from the molecular network of Myoporeae. For each specimen in this study, the total amount of metabolites of a corresponding chemical family is displayed within the heatmap. Left: The nuclear ribosomal DNA phylogeny on the left side presents the major phylogenetic clades. Top: As depicted on top of the heatmap, the chemical information underwent hierarchical clustering according to the given phylogeny to reveal chemo‐evolutionary patterns among subsets of chemical families, defined as heatmap metabolic clusters (HMCs) I–X. Bottom: Selected chemical classifications generated by NAP‐based dereplication are displayed below in blue, while the presence of level 1 identification (m/z, retention time, and MS2 match) in a particular chemical family is shown in gold. Right: Functional annotations including the presence of leaf resin and hairiness, pollination, antibacterial activity, traditional medicinal usage, and biogeographical species distribution information are displayed on the right side. A summary of legends is also included to the right of the figure.
Figure S5. Comparison of species distribution between members of TMC A‐ and B‐related chemistry. Species distributions for members of tanglegram metabolite clusters A (A1–A6) and B (B1 and B2). Widespread species (those with broad distributions spanning more than one biome) have been excluded from maps. Species assessed as widespread were identified in clusters A1 (Eremophila bignoniiflora, Eremophila alternifolia, Eremophila deserti), A2 (Myoporum montanum, Eremophila latrobei subsp. glabra, Eremophila deserti), A5 (Myoporum acuminatum, Eremophila longifolia), and B2 (Eremophila mitchellii). Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 29 June 2020.
Figure S6. Geographic distributions of two widespread Eremophila species. Eremophila alternifolia and Eremophila duttonii specimen locations are marked with red boxes. Data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020.
Figure S7. Molecular network analysis of Eremophila deserti. Global molecular network representation of metabolite distribution from three E. deserti specimens. Highlighted in yellow are putative metabolites that are shared by at least two specimen of E. deserti. Metabolites found specifically in one of the specimens are marked in blue: E. deserti (MJB2384), red: E. deserti (RMF228), and green: E. deserti (RMF204). On each node in the network, the total number of specimens that share this specific metabolite is displayed. Indicated on the right side is the geographic distribution of the observed E. deserti specimen in this study, with corresponding sampling locations marked with colored boxes. Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020. Maps of Australian annual mean temperature, annual mean precipitation, and annual mean solar radiation were downloaded as layers for reference from the Atlas of Living Australia (ala.org.au).
Figure S8. Spectral comparison of three Eremophila deserti specimens. Spectral representation of base peak chromatograms for three E. deserti specimens that display interspecific chemical variation. Selected compounds are highlighted and chemical annotations are stated below. An extracted ion chromatogram of specimen RMF204 is also shown to show the complete lack of flavonoid compounds present in specimen MJB2384.
Figure S9. Species distributions for members of Myoporeae phylogenetic clades A–H. In this representation, widespread species (those with broad distributions spanning more than one biome) have been excluded from maps. Species assessed as widespread were identified in phylogenetic clades A (Myoporum acuminatum, Myoporum montanum) D (Eremophila bignoniiflora, Eremophila deserti, Eremophila polyclada), G (Eremophila alternifolia), and H (Eremophila latrobei subsp. glabra, Eremophila longifolia, Eremophila maculata subsp. maculata, Eremophila mitchellii). Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020.
Figure S10. Climatic information about Australia. Maps of Australian annual mean temperature, annual mean precipitation, annual mean solar radiation, and annual mean relative humidity, as well as major classes of the Koppen Climate Classification were downloaded as layers for reference for the species distribution maps in this study from the Atlas of Living Australia (ala.org.au).
Figure S11. Species distributions for members of Myoporeae phylogenetic clade H, subclades H1–H14. In this representation, widespread species (those with broad distributions spanning more than one biome) have been excluded from maps. Species assessed as widespread were identified in phylogenetic subclades H2 (Eremophila mitchellii), H5 (contains only Eremophila longifolia, so no distribution map was included), H8 (Eremophila maculata subsp. maculata), and H10 (Eremophila latrobei subsp. glabra). Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020.
Figure S12. Continuous heatmap analysis of chemo‐evolutionary patterns of serrulatane and viscidane diterpenoids in Myoporeae. A heatmap analysis was conducted to put chemical information into an evolutionary context. For that, information about the 30 chemical families derived from the molecular network of Myoporeae which have been found to be associated with serrulatane and viscidane diterpene chemistry was compiled. For each specimen in this study, the total amount of metabolites of a corresponding chemical family is displayed within the heatmap. Left: The nuclear ribosomal DNA phylogeny on the left side presents the major phylogenetic clades. Top: As depicted on top of the heatmap, the chemical information was subjected to hierarchical clustering according to the given phylogeny to reveal chemo‐evolutionary patterns. Bottom: Selected chemical classifications generated by NAP‐based dereplication are displayed below in blue, while the presence of level 1 identification (m/z, retention time, and MS2 match) in a particular chemical family is shown in gold. Right: Functional annotations including the presence of leaf resin and hairiness, pollination, antibacterial activity, traditional medicinal usage, and biogeographical species distribution information, as well as tanglegram metabolic cluster (TMC) associations, are displayed on the right side. A summary of legends is also included to the right of the figure.
Figure S13. Annotation of substructural motif ‘Eremophila motif 440’. Annotation of the unknown substructural motif ‘Eremophila motif 440’ was conducted using the combined information from the biochemical reaction prediction tool MetWork and the in silico MS2 spectrum prediction tool CFM‐ID. (a) The spectral representation of the studied MS2LDA motif indicates the probabilities of presence and absence of distinct fragments, which have been annotated using in silico spectrum prediction by CFM‐ID on serrulatane diterpenoid structures found to be associated with ‘Eremophila motif 440’, such as KU006‐14. Based on these structural annotations a dihydroxy serrulatane acid bicyclic core was proposed as the substructure underlying this motif. (b) MetWork‐based prediction of biochemical and structural reactions joining serrulatane diterpenoid features present in chemical family 10 that are associated with ‘Eremophila motif 440’. The predictions indicate the loss of the motif upon dehydroxylation at the aromatic ring, underlying these specific modifications as the inherent motif characteristics. Nodes highlighted in light green/yellow correspond to prior dereplicated spectral features, while dark green displays unknown features that where structurally annotated by the MetWork analysis.
Figure S14. Spectral representation of MS2LDA‐predicted substructural motifs found in chemical families 10 and 22. Spectral information on the probabilities of mass fragments being present or absent within different substructural motifs was determined in chemical families 10 and 22 by the MS2LDA tool.
Figure S15. Phylogeny of tribe Myoporeae based on analysis of nuclear ribosomal DNA sequences. Bayesian inference 50% majority‐rule consensus tree, showing posterior probability values on each branch. Branches with values of <0.50 collapsed.
Figure S16. LC‐QTOF‐HRMS data quality control. The quality of the full spectral dataset used in this study was assessed by plotting all samples (red) together with the quality controls (blue) within a principal component analysis.
ACKNOWLEDGMENTS
This work was supported by the VILLUM Center for Plant Plasticity (VKR023054) (BLM), the European Research Council Advanced Grant (ERC‐2012‐ADG 20120314) (BLM), the Lundbeck Foundation (R223‐2016‐85, ‘Brewing diterpenoids’), the Cybec Foundation (Jim Ross Scholarship), and the Novo Nordisk Foundation Interdisciplinary Synergy (NNF 16OC0021616, ‘Desert‐loving therapeutics’) and Distinguished Investigator 2019 (NNF 0054563, ‘The Black Holes in the Plant Universe’) programs (BLM). We would like to thank Tanja Schuster for specimen collections and Bob Chinnock and Ron and Claire Dadd for access to their private garden collections. We thank the Australian National Botanic Garden, Canberra, the Australian Arid Lands Botanic Garden, Port Augusta, and the National Tropical Botanic Garden, Kauai for collection of garden specimens. We thank Elizabeth H. J. Neilson (UCPH, Denmark) for initiating the chemical profiling work of Eremophila species, Madeleine Ernst (SSI, Denmark) for the advice regarding molecular networking tools, Yong Zhao (UCPH, Denmark) and Laura Mikél Mc Nair (UCPH, Denmark) for providing reference compounds to establish the in‐house spectral library, and Dominik Merges (BiK‐F, Frankfurt) and the PLEN R‐club for additional advice regarding multivariate statistics.
DATA AVAILABILITY STATEMENT
All DNA sequences included in the phylogenetic analysis for this manuscript have been submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with accession numbers given in Table S1. The DNA sequence alignment and resulting tree have been submitted to TreeBase (https://www.treebase.org/treebase‐web/home.html) and are available for reviewer consideration at http://purl.org/phylo/treebase/phylows/study/TB2:S26197?x‐access‐code=8f551ae84c6f32323d73a86cc9e8d1bd&format=html.
Raw spectral data derived from the 291 specimens, additional quality checks, and blank samples are available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000086644.
This MASSIVE database repository also contains the metadata information used in this study (file name ‘Eremophila ChemoEvo metadata complete QC BLANK update.txt’ in the ‘metadata’ tab) as well as the MZmine2 output before (file name ‘20200220 MZMINE OUTPUT.csv’) and after normalization (‘20200220 MZMINE OUTPUT AFTER NORMALISATION.csv’) as well as spectral database files for in silico prediction used during the NAP workflow as supplementary files (‘20200222 Eremophila ISDB.tsv/csv’ within the ‘other’ tab found in ‘Browse Dataset Files’) for download. Metadata information is interpreted as follows: leaf resin: ‘0’ – absence, ‘1’ – presence; leaf hairiness: ‘0’ – absence, ‘1’ – presence; pollination: ‘0’ – insect, ‘1’ – bird; traditional use: ‘0’ – no documented use, ‘1’ – use is documented; antibacterial activity: ‘0’ – tested absence of activity, ‘1’ tested presence of activity (corresponding references are found in Table S1 of the manuscript); biogeographical distribution: ‘0’ – widespread, ‘1’ – Eremean, ‘2’ – Southwestern temperate, ‘3’ – Southeastern temperate, ‘5’ – non‐Australian; general phylogenetic clades: ‘o’ – outgroup, ‘ag’ – clades A to G, ‘h3’ – clade H3, ‘h10’ – clade H10, ‘h11’ – clade H11, ‘h12’ – clade H12, ‘h14’ – clade H14, and ‘h’ – remaining clade H‐associated taxa; sample weight: weight in mg of the plant material used for extraction per sample. Described data files are also found within the digital supplement in addition to all relevant tables from this study in machine‐readable formats as well as compiled in one Excel sheet. In addition, all dendrograms used to generate tanglegrams (Newick format) as well as the underlying data matrices (tab separated) for the metabolic cluster analysis shown in Figure 3 and Figure S3 are provided within the digital supplement. Thereby individual spectral features are often indicated as follows: ‘[compound ID] [mass] [retention time]’.
The molecular networking job can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f74d3374a7ba43eb8d58e5d2a312bf33.
The NAP job can be publicly accessed at https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=5980fea229b9427bb610a8e7d6c25827. The MS2LDA job can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8082c8133d1843c39af42164157ab043.
Spectral information derived from the 76 reference compounds used in this study is available at the GNPS spectral library under the CCMSLIB IDs given in Table S3.
Underlying data for geographic distribution maps are available for:
Figure S5 TMC A1: https://doi.ala.org.au/doi/10.26197/ala.337ac334‐016c‐4ad3‐a8dd‐f9ce79aef8a7
TMC A2: https://doi.ala.org.au/doi/10.26197/ala.4693c8c5‐cad1‐4824‐ba5f‐c4d25aa38686
TMC A3: https://doi.ala.org.au/doi/10.26197/ala.a026ad9e‐56a3‐482b‐a2f7‐fa2201b9c3b2
TMC A4: https://doi.ala.org.au/doi/10.26197/ala.edb84e7b‐9e4c‐479e‐b581‐300eb4d27bcc
TMC A5: https://doi.ala.org.au/doi/10.26197/ala.af1cf180‐7c18‐42eb‐93b2‐6dbe8b27d399
TMC A6: https://doi.ala.org.au/doi/10.26197/ala.fdcb84c8‐5db6‐4fea‐8f93‐2101fb0d2af0
TMC B1: https://doi.ala.org.au/doi/10.26197/ala.6ce9b49c‐d67f‐4242‐83ef‐495868db9800
TMC B2: https://doi.ala.org.au/doi/10.26197/ala.2a8e032c‐bbb3‐4bd1‐936a‐2ca83269b1a8
Figure S6 Eremophila alternifolia: https://doi.ala.org.au/doi/10.26197/ala.cacf63e9‐5681‐4581‐af10‐d1c6f4799143
Eremophila duttonii: https://doi.ala.org.au/doi/10.26197/ala.a6acf423‐00b3‐41f4‐8d5c‐dddcc6553f54
Figure S7 Eremophila deserti: https://doi.ala.org.au/doi/10.26197/ala.f2d34675‐c1ae‐4a81‐adf9‐fe03e34cd388
Figure S9 Clade A: https://doi.ala.org.au/doi/10.26197/ala.3e501c4c‐da8547e4‐985f‐2af4d6365766
Clade B: https://doi.ala.org.au/doi/10.26197/ala.3e0044f8‐5383‐4106‐8fb4‐ae0a3a2be0a6
Clade C: https://doi.ala.org.au/doi/10.26197/ala.2528df04‐a91d‐42b8‐88f0‐6f66bfca3f61
Clade D: https://doi.ala.org.au/doi/10.26197/ala.f82d15d5‐f54a‐431b‐b690‐c193553d55c4
Clade E: https://doi.ala.org.au/doi/10.26197/ala.6cc5e4e9‐8560‐4d31‐8550‐6576c0b3b2b0
Clade F: https://doi.ala.org.au/doi/10.26197/ala.99838369‐78e7‐4de5‐aac4‐20feeea4e748
Clade G: https://doi.ala.org.au/doi/10.26197/ala.2d5c0411‐960b‐4d87‐9e98‐186032eb7ccb
Clade H: https://doi.ala.org.au/doi/10.26197/ala.2d2f885c‐ff52‐4b89‐aa00‐b121ab789a66
Figure S11 Clade H1: https://doi.ala.org.au/doi/10.26197/ala.ff62d761‐f339‐4933‐88ae‐0087fdaf84a0
Clade H2: https://doi.ala.org.au/doi/10.26197/ala.e00c3310‐66f5‐42a4‐9df8‐52a5e0cc50ab
Clade H3: https://doi.ala.org.au/doi/10.26197/ala.53afe67f‐cccf‐4dfb‐98cd‐ce3a935bdfb7
Clade H4: https://doi.ala.org.au/doi/10.26197/ala.6a389f01‐9668‐4de5‐af9e‐f83f75478cb8
Clade H6: https://doi.ala.org.au/doi/10.26197/ala.ddf5f8b9‐87a9‐4b77‐b59d‐389ae763231b
Clade H7: https://doi.ala.org.au/doi/10.26197/ala.ef9633ba‐4eef‐43c5‐951b‐f8a065c1033e
Clade H8: https://doi.ala.org.au/doi/10.26197/ala.5fdea1f0‐8854‐4fc0‐b26e‐22cec0d4242c
Clade H9: https://doi.ala.org.au/doi/10.26197/ala.6a655fd9‐a8ee‐4174‐8ffa‐ce7cab21dc12
Clade H10: https://doi.ala.org.au/doi/10.26197/ala.02071a0a‐7523‐4380‐96ac‐9485dc3388db
Clade H11: https://doi.ala.org.au/doi/10.26197/ala.35441d4d‐99c0‐43fa‐a4ee‐eea1bc6160c4
Clade H12: https://doi.ala.org.au/doi/10.26197/ala.c18cf6c9‐eba3‐48ac‐82a5‐13af5983f892
Clade H13: https://doi.ala.org.au/doi/10.26197/ala.6bea9d4f‐7c52‐43c6‐983a‐359e60d01832
Clade H14: https://doi.ala.org.au/doi/10.26197/ala.19793839‐fd96‐4dda‐b552‐d157f1e77af4
REFERENCES
- Afzan, A. , Bréant, L. , Bellstedt, D.U. , Grant, J.R. , Queiroz, E.F. , Wolfender, J.‐L. et al. (2019) Can biochemical phenotype, obtained from herbarium samples, help taxonomic decisions? – A case study using Gentianaceae. Taxon, 68(4), 771–782. [Google Scholar]
- Algreiby, A.A. , Hammer, K.A. , Durmic, Z. , Vercoe, P. & Flematti, G.R. (2018) Antibacterial compounds from the Australian native plant Eremophila glabra . Fitoterapia, 126, 45–52. [DOI] [PubMed] [Google Scholar]
- Aminimoghadamfarouj, N. & Nematollahi, A. (2017) Structure elucidation and botanical characterization of diterpenes from a specific type of bee glue. Molecules, 22, 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J. (2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecology, 26(1), 32–46. [Google Scholar]
- Anderson, M.J. & Walsh, D.C.I. (2013) PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecological Monographs, 83(4), 557–574. [Google Scholar]
- Aron, A.T. , Gentry, E.C. , McPhail, K.L. , Nothias, L.‐F. , Nothias‐Esposito, M. , Bouslimani, A. et al. (2020) Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols, 15, 1954–1991. [DOI] [PubMed] [Google Scholar]
- Barnes, E.C. , Kavanagh, A.M. , Ramu, S. , Blaskovich, M.A. , Cooper, M.A. & Davis, R.A. (2013) Antibacterial serrulatane diterpenes from the Australian native plant Eremophila microtheca . Phytochemistry, 93, 162–169. [DOI] [PubMed] [Google Scholar]
- Barr, A. (1988) Traditional bush medicines: an aboriginal pharmacopoeia. Richmond, Vic., Australia: Greenhouse Publications. [Google Scholar]
- Barr, A. , Chapman, J. , Smith, N. , Wightman, G. & Knight, T. (1993) Traditional aboriginal medicines in the Northern Territory of Australia by Aboriginal Communities of the Northern Territory. (ed Conservation Commission of the Northern Territory) (Darwin, 1993). Darwin, Australia: Conservation Commission of the Northern Territory of Australia. [Google Scholar]
- Beauxis, Y. & Genta‐Jouve, G. (2019) MetWork: a web server for natural products anticipation. Bioinformatics, 35(10), 1795–1796. [DOI] [PubMed] [Google Scholar]
- Becerra, J.X. (2007) The impact of herbivore‐plant coevolution on plant community structure. Proceedings of the National Academy of Sciences of the United States of America, 104, 7483–7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop Museum . (2020) Hawaiian ethnobotany online database 2020. Available at: http://data.bishopmuseum.org/ethnobotanydb/ethnobotany.php?b=d&ID=naio [Accessed 23 April 2020].
- Biva, I.J. , Ndi, C.P. , Griesser, H.J. & Semple, S.J. (2016) Antibacterial constituents of Eremophila alternifolia: an Australian aboriginal traditional medicinal plant. Journal of Ethnopharmacology, 182, 1–9. [DOI] [PubMed] [Google Scholar]
- Biva, I.J. , Ndi, C.P. , Semple, S.J. & Griesser, H.J. (2019) Antibacterial performance of terpenoids from the Australian plant Eremophila lucida . Antibiotics, 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, S.E. (1975) Taxonomic studies in the Myoporaceae. PhD Thesis, The University of New England. [Google Scholar]
- Byrne, M. , Yeates, D.K. , Joseph, L. , Kearney, M. , Bowler, J. , Williams, M.A.J. et al. (2008) Birth of a biome: Insights into the assembly and maintenance of the Australian arid zone biota. Molecular Ecology, 17, 4398–4417. [DOI] [PubMed] [Google Scholar]
- Cassis, G. & Symonds, C. (2008) Systematics, biogeography and host associations of the lace bug genus Inoma (Hemiptera: Heteroptera: Tingidae). Acta Entomologica Musei Nationalis Pragae, 48, 433–484. [Google Scholar]
- Chinnock, R.J. (2007) Eremophila and allied genera. A monograph of the Myoporaceae. Kenthurst, NSW, Australia: Rosenberg Publishing. [Google Scholar]
- Clarke, P.A. (2013) The aboriginal ethnobotany of the Adelaide Region, South Australia. Transactions of the Royal Society of South Australia, 137(1), 97–126. [Google Scholar]
- Clarke, P.A. (2014) Discovering aboriginal plant use: the journeys of an Australian Anthropologist. Kenthurst, NSW, Australia: Rosenberg Publishing. [Google Scholar]
- Clealand, J.B. & Tindale, N.B. (1959) The native names and uses of plants at Haast Bluff, Central Australia. Transactions and proceedings of the Royal Society of South Australia, 82, 123. [Google Scholar]
- Crisp, M.D. & Cook, L.G. (2013) How was the Australian flora assembled over the last 65 million years? A molecular phylogenetic perspective. Annual Review of Ecology, Evolution, and Systematics, 44(1), 303–324. [Google Scholar]
- Crisp, M. , Cook, L. & Steane, D. (2004) Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present–day communities? Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 359(1450), 1551–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, G.M. , Mulham, W. , Milthorpe, P. & Leigh, J. (1981) Plants of western New South Wales. Sydney, Australia: NSW Government Printer. [Google Scholar]
- Da Silva, R.R. , Wang, M. , Nothias, L.‐F. , van der Hooft, J.J.J. , Caraballo‐Rodríguez, A.M. , Fox, E. et al. (2018) Propagating annotations of molecular networks using in silico fragmentation. PLOS Computational Biology, 14(4), e1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou‐Feunang, Y. , Pon, A. , Karu, N. , Zheng, J. , Li, C. , Arndt, D. et al. (2019) CFM‐ID 3.0: significantly improved ESI‐MS/MS prediction and compound identification. Metabolites, 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobler, S. , Petschenka, G. & Pankoke, H. (2011) Coping with toxic plant compounds – the insect’s perspective on iridoid glycosides and cardenolides. Phytochemistry, 72(13), 1593–1604. [DOI] [PubMed] [Google Scholar]
- Dudek, B. , Warskulat, A.C. & Schneider, B. (2016) The occurrence of flavonoids and related compounds in flower sections of Papaver nudicaule . Plants, 5, 93–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop, K. , Fleischauer, M. , Ludwig, M. , Aksenov, A.A. , Melnik, A.V. , Meusel, M. et al. (2019) SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nature Methods, 16, 299–302. [DOI] [PubMed] [Google Scholar]
- Dührkop, K. , Nothias, L.‐F. , Fleischauer, M. , Reher, R. , Ludwig, M. , Hoffmann, M.A. et al. (2021) Systematic classification of unknown metabolites using high‐resolution fragmentation mass spectra. Nature Biotechnology, 39, 462–471. 10.1038/s41587-020-0740-8 [DOI] [PubMed] [Google Scholar]
- Duhrkop, K. , Shen, H. , Meusel, M. , Rousu, J. & Böcker, S. (2015) Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proceedings of the National Academy of Sciences, 112(41), 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebach, M.C. & Murphy, D.J. (2020) Carving up Australia's arid zone: a review of the bioregionalisation of the Eremaean and Eyrean biogeographic regions. Australian Journal of Botany, 68(3), 229. [Google Scholar]
- Engels, B. , Dahm, P. & Jennewein, S. (2008) Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metabolic Engineering, 10, 201–206. [DOI] [PubMed] [Google Scholar]
- Ernst, M. , Kang, K.B. , Caraballo‐Rodríguez, A.M. , Nothias, L.‐F. , Wandy, J. , Chen, C. et al. (2019a) MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites, 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, M. , Nothias, L.F. , van der Hooft, J. , Silva, R.R. , Saslis‐Lagoudakis, C.H. , Grace, O.M. et al. (2019b) Assessing specialized metabolite diversity in the cosmopolitan plant genus Euphorbia L. Frontiers in Plant Science, 10, 846. 10.3389/fpls.2019.00846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L. , Briscoe, J. , Baker, E. , Barr, A. , Locher, C. , Muir, K. et al. (2010) Plants for people: laboratory study report. Desert Knowledge CRC Report. Alice Springs, Australia: Desert Knowledge Cooperative Research Centre. [Google Scholar]
- Fowler, R.M. , McLay, T.G.B. , Schuster, T.M. , Buirchell, B.J. , Murphy, D.J. & Bayly, M.J. (2020) Plastid phylogenomic analysis of tribe Myoporeae (Scrophulariaceae). Plant Systematics and Evolution, 306 (3). [Google Scholar]
- Fowler, R.M. , Murphy, D.J. , McLay, T.G.B. , Buirchell, B.J. , Chinnock, R.J. & Bayly, M.J. (2021) Molecular phylogeny of tribe Myoporeae (Scrophulariaceae) using nuclear ribosomal DNA : generic relationships and evidence for major clades. Taxon, 70(3), 570–588. [Google Scholar]
- Fox Ramos, A.E. , Evanno, L. , Poupon, E. , Champy, P. & Beniddir, M.A. (2019) Natural products targeting strategies involving molecular networking: different manners, one goal. Natural Product Reports, 36, 960–980. [DOI] [PubMed] [Google Scholar]
- Galappathie, S. et al. (2017) Antibacterial nerol cinnamates from the Australian plant Eremophila longifolia . Journal of Natural Products, 80, 1178–1181. [DOI] [PubMed] [Google Scholar]
- Galili, T. (2015) dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics, 31, 3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gericke, O. , Hansen, N.L. , Pedersen, G.B. , Kjaerulff, L. , Luo, D. , Staerk, D. et al. (2020) Nerylneryl diphosphate is the precursor of serrulatane, viscidane and cembranetype diterpenoids in Eremophila species. BMC Plant Biology, 20, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisalberti, E.L. , Jefferies, P.P. & Sheppard, P. (1975) A new class of diterpenes from Eremophila decipien . Tetrahedron Letters, 16(22–23), 1775–1778. [Google Scholar]
- Guijas, C. , Montenegro‐Burke, J.R. , Domingo‐Almenara, X. , Palermo, A. , Warth, B. , Hermann, G. et al. (2018) METLIN: a technology platform for identifying knowns and unknowns. Analytical Chemistry, 90, 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllenhaal, C. , Kadushin, M.R. , Southavong, B. , Sydara, K. , Bouamanivong, S. , Xaiveu, M. et al. (2012) Ethnobotanical approach versus random approach in the search for new bioactive compounds: support of a hypothesis. Pharmaceutical Biology, 50, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, A.L. , Edrada‐Ebel, R. & Quinn, R.J. (2015) The re‐emergence of natural products for drug discovery in the genomics era. Nature Reviews Drug Discovery, 14, 111–129. [DOI] [PubMed] [Google Scholar]
- van der Hooft, J.J.J. , Wandy, J. , Barrett, M.P. , Burgess, K.E.V. & Rogers, S. (2016) Topic modeling for untargeted substructure exploration in metabolomics. Proceedings of the National Academy of Sciences of the United States of America, 113(48), 13738–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai, H. , Arita, M. , Kanaya, S. , Nihei, Y. , Ikeda, T. , Suwa, K. et al. (2010) MassBank: a public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry, 45, 703–714. [DOI] [PubMed] [Google Scholar]
- Huber, F. , Ridder, L. , Verhoeven, S. , Spaaks, J.H. , Diblen, F. , Rogers, S. et al. (2021) Spec2Vec: improved mass spectral similarity scoring through learning of structural relationships. PLoS Computational Biology, 17, e1008724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, J. (1987) Bush food: aboriginal food and herbal medicine (Weldons, McMahons Point, N.S.W., Australia, 1987). McMahons Point, Australia: Weldons. [Google Scholar]
- Kang, K.B. , Ernst, M. , Hooft, J.J.J. , Silva, R.R. , Park, J. , Medema, M.H. et al. (2019) Comprehensive mass spectrometry‐guided phenotyping of plant specialized metabolites reveals metabolic diversity in the cosmopolitan plant family Rhamnaceae. The Plant Journal, 98, 1134–1144. [DOI] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. et al. (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, A. & Kalske, A. (2018) Plant secondary metabolite diversity and species interactions. Annual Review of Ecology, Evolution, and Systematics, 49, 115–138. [Google Scholar]
- Kjaerulff, L. , Jensen, A.B.J. , Ndi, C. , Semple, S. , Møller, B.L. & Staerk, D. (2020) Isolation, structure elucidation and PTP1B inhibitory activity of serrulatane diterpenoids from the roots of Myoporum insulare . Phytochemistry Letters, 39, 49–56. [Google Scholar]
- Kracht, O.N. , Ammann, A.‐C. , Stockmann, J. , Wibberg, D. , Kalinowski, J. , Piotrowski, M. et al. (2017) Transcriptome profiling of the Australian arid‐land plant Eremophila serrulata (A.DC.) Druce (Scrophulariaceae) for the identification of monoterpene synthases. Phytochemistry, 136, 15–22. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Duffy, S. , Avery, V.M. , Carroll, A.R. & Davis, R.A. (2018) Microthecaline A, a quinoline serrulatane alkaloid from the roots of the Australian desert plant Eremophila microtheca . Journal of Natural Products, 81, 1079–1083. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Duffy, S. , Avery, V.M. & Davis, R.A. (2017) Synthesis of antimalarial amide analogues based on the plant serrulatane diterpenoid 3,7,8‐trihydroxyserrulat‐14‐en‐19‐oic acid. Bioorganic and Medicinal Chemistry Letters, 27, 4091–4095. [DOI] [PubMed] [Google Scholar]
- Langenheim, J.H. (2004) Plant resins: chemistry, evolution, ecology, and ethnobotany. Annals of Botany, 93, 784–785. [Google Scholar]
- Lans, C.A. (2006) Ethnomedicines used in Trinidad and Tobago for urinary problems and diabetes mellitus. Journal of Ethnobiology and Ethnomedicine, 2, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak, E. & McCarthy, T. (1983) Australian medicinal plants. Sydney, Australia: Methuen Australia. ISBN: 0454004389. [Google Scholar]
- Latz, P.K. (1982) Bushfires and Bushtucker PhD thesis (The University of New England, 1982), 244.
- Latz, P. , Green, J. ; Institute for Aboriginal Development . (1995) Bushfires & bushtucker: aboriginal plant use in Central Australia. Alice Springs, N.T: I.A.D. Press. [Google Scholar]
- Liu, Q. , Harrington, D. , Kohen, J.L. , Vemulpad, S. & Jamie, J.F. (2006) Bactericidal and cyclooxygenase inhibitory diterpenes from Eremophila sturtii . Phytochemistry, 67, 1256–1261. [DOI] [PubMed] [Google Scholar]
- Lyddiard, D. & Greatrex, B.W. (2018) Serrulatic acid diastereomers identified from an antibacterial survey of Eremophila. Fitoterapia, 126, 29–34. [DOI] [PubMed] [Google Scholar]
- Mafu, S. & Zerbe, P. (2018) Plant diterpenoid metabolism for manufacturing the biopharmaceuticals of tomorrow: prospects and challenges. Phytochemistry Reviews, 17, 113–130. [Google Scholar]
- Meggitt, M. (1962) Desert people: a study of the Walbiri aborigines of Central Australia. Melbourne, Australia: Angus & Robertson. [Google Scholar]
- Mijajlovic, S. , Smith, J. , Watson, K. , Parsons, P. & Jones, G. (2006) Traditional Australian medicinal plants: screening for activity against human cancer cell lines. Journal of the Australian Traditional‐Medicine Society, 12, 129–132. [Google Scholar]
- Mon, H.H. et al. (2015) Serrulatane diterpenoid from Eremophila neglecta exhibits bacterial biofilm dispersion and inhibits release of pro‐inflammatory cytokines from activated macrophages. Journal of Natural Products, 78, 3031–3040. [DOI] [PubMed] [Google Scholar]
- Moorthy, A.S. , Wallace, W.E. , Kearsley, A.J. , Tchekhovskoi, D.V. & Stein, S.E. (2017) Combining fragment‐ion and neutral‐loss matching during mass spectral library searching: a new general purpose algorithm applicable to illicit drug identification. Analytical Chemistry, 89, 13261–13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D.J. , Ebach, M.C. , Miller, J.T. , Laffan, S.W. , Cassis, G. , Ung, V. et al. (2019) Do phytogeographic patterns reveal biomes or biotic regions? Cladistics, 35(6), 654–670. [DOI] [PubMed] [Google Scholar]
- Ndi, C.P. , Semple, S.J. , Griesser, H.J. & Barton, M.D. (2007a) Antimicrobial activity of some Australian plant species from the genus Eremophila. Journal of Basic Microbiology, 47, 158–164. [DOI] [PubMed] [Google Scholar]
- Ndi, C.P. , Semple, S.J. , Griesser, H.J. , Pyke, S.M. & Barton, M.D. (2007b) Antimicrobial compounds from the australian desert plant Eremophila neglecta . Journal of Natural Products, 70, 1439–1443. [DOI] [PubMed] [Google Scholar]
- Ndi, C.P. , Semple, S.J. , Griesser, H.J. , Pyke, S.M. & Barton, M.D. (2007c) Antimicrobial compounds from Eremophila serrulata . Phytochemistry, 68(21), 2684–2690. [DOI] [PubMed] [Google Scholar]
- Ngo, L.T. , Okogun, J.I. & Folk, W.R. (2013) 21st Century natural product research and drug development and traditional medicines. Natural Product Reports, 30, 584–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L.T. , Schmidt, H.A. , Von Haeseler, A. & Minh, B.Q. (2015) IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias, L.‐F. , Nothias‐Esposito, M. , da Silva, R. , Wang, M. , Protsyuk, I. , Zhang, Z. et al. (2018) Bioactivity‐based molecular networking for the discovery of drug leads in natural product bioassay‐guided fractionation. Journal of natural products, 81, 758–767. [DOI] [PubMed] [Google Scholar]
- Nothias, L.F. , Petras, D. , Schmid, R. , Dührkop, K. , Rainer, J. , Sarvepalli, A. et al. (2020) Feature‐based molecular networking in the GNPS analysis environment. Nature Methods, 17, 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell, J.F. , Latz, P.K. & Peggy, B. (1983) Traditional and modern plant use among the Alyawara of central Australia. Economic Botany, 37(1), 80–109. [Google Scholar]
- Ogbourne, S.M. & Parsons, P.G. (2014) The value of nature’s natural product library for the discovery of new chemical entities: the discovery of ingenol mebutate. Fitoterapia, 98, 36–44. [DOI] [PubMed] [Google Scholar]
- Oksanen, J. et al. (2019) Vegan: community ecology package. R package version 2.5‐6.
- Olivon, F. , Allard, P.‐M. , Koval, A. , Righi, D. , Genta‐Jouve, G. , Neyts, J. et al. (2017) Bioactive natural products prioritization using massive multi‐informational molecular networks. ACS Chemical Biology, 12, 2644–2651. [DOI] [PubMed] [Google Scholar]
- Olivon, F. , Apel, C. , Retailleau, P. , Allard, P.M. , Wolfender, J.L. , Touboul, D. et al. (2018) Searching for original natural products by molecular networking: detection, isolation and total synthesis of chloroaustralasines. Organic Chemistry Frontiers, 5(14), 2171–2178. [Google Scholar]
- Palombo, E.A. & Semple, S.J. (2001) Antibacterial activity of traditional Australian medicinal plants. Journal of Ethnopharmacology, 77, 151–157. [DOI] [PubMed] [Google Scholar]
- Palombo, E.A. & Semple, S.J. (2002) Antibacterial activity of Australian plant extracts against methicillin‐resistant Staphylococcus aureus (MRSA) and vancomycin‐resistant enterococci (VRE). Journal of Basic Microbiology, 42(6), 444–448. [DOI] [PubMed] [Google Scholar]
- Panche, A.N. , Diwan, A.D. & Chandra, S.R. (2016) Flavonoids: an overview. Journal of Nutritional Science, 5, 10.1017/jns.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateraki, I. , Andersen‐Ranberg, J. , Jensen, N.B. , Wubshet, S.G. , Heskes, A.M. , Forman, V. et al. (2017) Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii . eLife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, H.A. et al. (2020) PTP1B‐inhibiting branched‐chain fatty acid dimers from Eremophila oppositifolia subsp. angustifolia identified by high‐resolution PTP1B inhibition profiling and HPLC‐PDA‐HRMS‐SPE‐NMR analysis. Journal of Natural Products, 83, 1598–1610. [DOI] [PubMed] [Google Scholar]
- Pluskal, T. , Castillo, S. , Villar‐Briones, A. & Oresic, M. (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry‐based molecular profile data. BMC Bioinformatics, 11(1), 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. , Drummond, A.J. , Xie, D. , Baele, G. & Suchard, M.A. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, L.A. , Dyer, L.A. , Forister, M.L. , Smilanich, A.M. , Dodson, C.D. , Leonard, M.D. et al. (2015) Phytochemical diversity drives plant‐insect community diversity. Proceedings of the National Academy of Sciences of the United States of America, 112, 10973–10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, G. (1993) A review of the use of Eremophila (Myoporaceae) by Australian Aborigines. Journal of the Adelaide Botanic Garden, 15, 101–107. [Google Scholar]
- Richmond, G.S. & Ghisalberti, E.L. (1994) The Australian Desert Shrub Eremophila (Myoporaceae): medicinal, cultural, horticultural and phytochemical uses. Botany‐Botanique, 48, 35–59. [Google Scholar]
- Richmond, G.S. & Ghisalberti, E.L. (1995) Cultural, food, medicinal uses and potential applications of Myoporum species (Myoporaceae). Economic Botany, 49, 276–285. [Google Scholar]
- Roberts, D.W. (2019) labdsv: ordination and multivariate analysis for ecology. R package version 2.0‐(2019).
- Rogers, K. , Fong, W. , Redburn, J. & Griffiths, L. (2002) Fluorescence detection of plant extracts that affect neuronal voltage‐gated Ca2+ channels. European Journal of Pharmaceutical Sciences, 15, 321–330. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. , Teslenko, M. , van der Mark, P. , Ayres, D.L. , Darling, A. , Höhna, S. et al. (2012) Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadgrove, N.J. et al. (2016) The iridoid myodesert‐1‐ene and elemol/eudesmol are found in distinct chemotypes of the Australian aboriginal medicinal plant Eremophila dalyana (Scrophulariaceae). Natural Product Communications, 83, 1598–1610. [PubMed] [Google Scholar]
- Sadgrove, N.J. & Jones, G.L. (2013) A possible role of partially pyrolysed essential oils in Australian Aboriginal traditional ceremonial and medicinal smoking applications of Eremophila longifolia (R. Br.) F. Muell (Scrophulariaceae). Journal of Ethnopharmacology, 147, 638–644. [DOI] [PubMed] [Google Scholar]
- Sadgrove, N.J. , Padilla‐González, G.F. , Green, A. , Langat, M.K. , Mas‐Claret, E. , Lyddiard, D. et al. (2021) The diversity of volatile compounds in Australia’s semi‐desert genus Eremophila (Scrophulariaceae). Plants, 10, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama, M.T.I. (2017) Antimicrobial activity of essential oil of Myoporum acuminatum R.Br fruits, cultivated in Libya. Journal of Essential Oil Bearing Plants, 20(1), 233–239. [Google Scholar]
- Schuster, T.M. , Setaro, S.D. , Tibbits, J.F.G. , Batty, E.L. , Fowler, R.M. , McLay, T.G.B. et al. (2018) Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS One, 13, e0195034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secretariat of the Convention on Biological Diversity (2011) Nagoya protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization to the convention on biological diversity. Technical Report (Montreal, 2011), 1–15.
- Semple, S.J. , Reynolds, G.D. , O’Leary, M.C. & Flower, R.L. (1998) Screening of Australian medicinal plants for antiviral activity. Journal of ethnopharmacology, 60, 163–172. [DOI] [PubMed] [Google Scholar]
- Shannon, P. (2003) Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, V. & Sarkar, I.N. (2013) Leveraging biodiversity knowledge for potential phyto‐therapeutic applications. Journal of the American Medical Informatics Association, 20, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, L.D. & McLay, T.G. (2011) Two micro‐scale protocols for the isolation of DNA from polysaccharide‐rich plant tissue. Journal of Plant Research, 124, 311–314. [DOI] [PubMed] [Google Scholar]
- Silberbauer, G.B. (1971) Ecology of the ernabella aboriginal community. Anthropological Forum, 3, 21–36. [Google Scholar]
- Singab, A.N. , Youssef, F.S. , Ashour, M.L. & Wink, M. (2013) The genus Eremophila (Scrophulariaceae): an ethnobotanical, biological and phytochemical review. Journal of Pharmacy and Pharmacology, 65, 1239–1279. [DOI] [PubMed] [Google Scholar]
- da Silva, R.R. , Dorrestein, P.C. & Quinn, R.A. (2015) Illuminating the dark matter in metabolomics. Proceedings of the National Academy of Sciences, 112(41), 12549–12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.E. , Tucker, D. , Watson, K. & Jones, G.L. (2007) Identification of antibacterial constituents from the indigenous Australian medicinal plant Eremophila duttonii F. Muell. (Myoporaceae). Journal of Ethnopharmacology, 112, 386–393. [DOI] [PubMed] [Google Scholar]
- Smith, N.M. (1991) Ethnobotanical field notes from the Northern Territory, Australia. Journal of the Adelaide Botanic Gardens, 14, 1–65. [Google Scholar]
- Spencer, B. & Gillen, F.J. (1969) The Northern Tribes of Central Australia. Oosterhout, The Netherlands: Anthropological Publications. [Google Scholar]
- Symonds, C.L. & Cassis, G. (2014) A new genus Ittolemma (Heteroptera: Tingidae) gen. nov. and three included species of hirsute lace bugs from temperate woodlands of southern Australia. Austral Entomology, 53, 380–390. [Google Scholar]
- Tahtah, Y. et al. (2016) High‐resolution PTP1B inhibition profiling combined with high‐performance liquid chromatography–high‐resolution mass spectrometry–solid‐phase extraction–nuclear magnetic resonance spectroscopy: proof‐of‐concept and antidiabetic constituents in crude extra. Fitoterapia, 110, 52–58. [DOI] [PubMed] [Google Scholar]
- Tindale, N.B. (1937) Vocabulary of Pitjandjarra, the Language of the Native of the great western Desert (Adelaide) 1931‐1937 . 138pp. Adelaide, Australia
- Tynan, B.J. (1979) Medical systems in conflict. A study of power. PhD thesis (The University of Sydney). [Google Scholar]
- Veneziani, R.C. , Ambrosio, S.R. , Martins, C.H. , Lemes, D.C. & Oliveira, L.C. (2017) Antibacterial potential of diterpenoids. Studies in Natural Products Chemistry, 54, 109–139. [Google Scholar]
- Wang, M. , Carver, J.J. , Phelan, V.V. , Sanchez, L.M. , Garg, N. , Peng, Y. et al. (2016) Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology, 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Naumann, U. , Eddelbuettel, D. , Wilshire, J. & Warton, D. (2020) mvabund: statistical methods for analysing multivariate abundance data. R package version 4.1.3.
- Warton, D.I. , Wright, S.T. & Wang, Y. (2012) Distance‐based multivariate analyses confound location and dispersion effects. Methods in Ecology and Evolution, 3(1), 89–101. [Google Scholar]
- Webb, L.J. (1948) Guide to the medicinal and poisonous plants of Queensland Bulletin No (ed Council for Scientific and Industrial Research). Melbourne: Government Printer. [Google Scholar]
- Wong, W. (1976) Some folk medicinal plants from Trinidad. Economic Botany, 30, 103–142. [Google Scholar]
- Woodworth, R. (1943) Economic plants of St. John, U.S. Virgin Islands. Botanical Museum Leaflets, Harvard University 11, 29–54. [Google Scholar]
- Wubshet, S.G. , Tahtah, Y. , Heskes, A.M. , Kongstad, K.T. , Pateraki, I. , Hamberger, B. et al. (2016) Identification of PTP1B and α‐glucosidase inhibitory serrulatanes from Eremophila spp. by combined use of dual high‐resolution PTP1B and α‐glucosidase inhibition profiling and HPLC‐HRMS‐SPE‐NMR. Journal of Natural Products, 79(4), 1063–1072. [DOI] [PubMed] [Google Scholar]
- Xing, S. , Hu, Y. , Yin, Z. , Liu, M. , Tang, X. , Fang, M. et al. (2020) Retrieving and utilizing hypothetical neutral losses from tandem mass spectra for spectral similarity analysis and unknown metabolite annotation. Analytical Chemistry, 92, 14476–14483. [DOI] [PubMed] [Google Scholar]
- Zaleta‐Pinet, D. , McCluskey, A. , Hall, S. , Brophy, J. , Ashhurst‐Smith, C. , Sakoff, J. et al. (2016) The use of the toxic plant Myoporum montanum in a traditional Australian aboriginal medicine. Australian Journal of Chemistry, 69(2), 161. [Google Scholar]
- Zhao, Y. , Kjaerulff, L. , Kongstad, K.T. , Heskes, A.M. , Møller, B.L. & Staerk, D. (2019) 2(5H)‐Furanone sesquiterpenes from Eremophila bignoniiflora: high‐resolution inhibition profiling and PTP1B inhibitory activity. Phytochemistry, 166, 112054. [DOI] [PubMed] [Google Scholar]
- Zola, N. & Gott, B. (1992) Koorie Plants Koorie People. Traditional Aboriginal Food, Fibre and Healing Plants of Victoria. (Koorie Heritage Trust, 1992).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Specimen details for all samples included in this study. Specimen information includes collection number, species name (authority), Eremophila section, collection location, herbarium voucher number, GenBank accession number, and collected metadata used in this study. Collecting numbers followed by * denote specimens grown in cultivation; where provenance information was known this was recorded in latitude/longitude. Collection initials refer to: RMF (Rachael Fowler), MJB (Michael Bayly), BB (Bevan Buirchell), TMS (Tanja Schuster), FTBG (Fairchild Tropical Botanic Garden), NTBG (National Tropical Botanical Garden). Published records of traditional medicinal use for each species are recorded where available (Barr, 1988; Barr et al., 1993; Bishop Museum, 2020; Bowen, 1975; Clarke, 2013; Clarke, 2014; Clealand and Tindale, 1959; Cunningham et al., 1981; Evans et al., 2010; Isaacs, 1987; Lans, 2006; Lassak and McCarthy, 1983; Latz, 1982; Latz and Green, 1995; Liu et al., 2006; Meggitt, 1962; Ndi et al., 2007b; O’Connell et al., 1983; Richmond, 1993; Richmond and Ghisalberti, 1995; Sadgrove et al., 2016; Sadgrove and Jones, 2013; Silberbauer, 1971; Smith, 1991; Spencer and Gillen, 1969; Tindale, 1937; Tynan, 1979; Webb, 1948; Wong, 1976; Woodworth; Zaleta‐Pinet et al., 2016). Evidence of antimicrobial activity against human bacterial pathogens (Gram‐positive bacteria) for each species is recorded where available (Algreiby et al., 2018; Aminimoghadamfarouj and Nematollahi, 2017; Barnes et al., 2013; Biva et al., 2016; Biva et al., 2019; Evans et al., 2010; Galappathie et al., 2017; Liu et al., 2006; Lyddiard and Greatrex, 2018; Ndi et al., 2007a, 2007b, 2007c; Salama, 2017; Smith et al., 2007). Metadata information is interpreted as follows: leaf resin: ‘0’ – absence, ‘1’ – presence; leaf hairiness: ‘0’ – absence, ‘1’ – presence; pollination: ‘0’ – insect, ‘1’ – bird; traditional use: ‘0’ – no documented use, ‘1’ – use is documented; antibacterial activity: ‘0’ – tested absence of activity, ‘1’ – tested presence of activity; biogeographical distribution: ‘0’ – widespread, ‘1’ – Eremean, ‘2’ – Southwestern temperate, ‘3’ – Southeastern temperate, ‘5’ –non‐Australian; phylogenetic clade association: ‘o’ – outgroup, ‘ag’ – clades A to G, ‘h3’ – clade H3, ‘h10’ – clade H10, ‘h11’ – clade H11, ‘h12’ – clade H12, ‘h14’ – clade H14, and ‘h’ – remaining clade H‐associated taxa.
Table S2. sirius‐based chemical classification of 10 singletons with the highest abundance. The table shows spectral information as well as the results of the chemical annotation of 10 singleton features found to have the highest abundance (peak signal intensity) in the Myoporeae spectral dataset. Chemical annotation was conducted here using sirius software and did not rely on feature‐based networking.
Table S3. List of reference compounds used as in‐house spectral database. Detailed spectrometric and structural information about all reference compounds used in this study.
Table S4. Detailed information for generated tanglegrams in this study. The table shows detailed information about common subbranches found in each tanglegram, which are indicated with a distinct taxon group ID and corresponding specimen ID tag for comparison. Further below are displayed the total number of taxa groups per tanglegram and the corresponding entanglement factor.
Table S5. Statistical analysis of individual heatmap metabolic cluster using general linear models. Generalized linear models were used to test for significant differences of functional annotations among the investigated specimens within a heatmap metabolic cluster (HMC) background. Models were generated for each HMC–attribute combination using the clogloc model, while excluding specimens with missing functional annotation. Each test was conducted using the anova.manyglm() function from the mvabund package. A P‐value of ≤0.05 was considered significant. ‘Res. Df’ – residual degrees of freedom; ‘Df.diff’ – model degrees of freedom; ‘Deviance’ – deviance of a fitted multivariate model object for abundance data.
Table S6. Chemical comparison of tanglegram metabolic cluster (TMC) A and B. Comparison of the chemical content between TMC A and B. The underlying metabolic cluster analysis is based on the 100 largest chemical families of the generated molecular network of Myoporeae. Numbers of specimens from major phylogenetic clades are shown with ‘O’: outgroup, ‘AG’: clades A to G, ‘H3’: clade H3, ‘H10’: clade H10, ‘H11’: clade H11, ‘H12’: clade H12, ‘H14’: clade H14, and ‘H’: remaining clade H‐associated taxa. Dominant chemical families are depicted for each TMC, with ‘CF80’: chemical families present in 80% of the specimens, ‘CF50’ and ‘CF30’: 50% and 30% abundancy of a given family among the given specimens, respectively. Different levels of predicted chemical classification and corresponding scores are shown for each chemical family. If present, level 1 identifications and their associated molecular structures are displayed.
Table S7. Multivariate statistical analysis involving the chemical space of Myoporeae. Permutational multivariate analysis of variance (permanova) was conducted involving all samples associated with a given functional annotation. Results of the Adonis2() and betadisper() functions are shown, which give information about statistical significance and data dispersion of tested attribute levels, respectively.
Table S8. Univariate tests and indicator species analysis of individual chemical families. To determine individual chemical families as overall significantly associated to a functional annotation, a univariate test was conducted using generalized linear models including all specimens with corresponding metadata to receive adjusted P‐values. An additional indicator species analysis using the indval() function was conducted to indicate the direction of annotation (level) for each chemical family.
Table S9. MZmine2 parameters used for raw chromatogram data processing. Detailed overview of parameters used for raw chromatogram data processing done in this study. This was carried out using the software MzMine2 (Pluskal et al., 2010).
Table S10. Molecular networking parameters. Detailed overview of parameters used for the feature‐based molecular networking approach presented in this study. The analysis was carried out using the Global Natural Products Social Molecular Networking (GNPS) toolbox (Wang et al., 2016).
Table S11. List of the 100 largest chemical families derived from the molecular network of Myoporeae. Detailed information about the largest 100 chemical families generated by the molecular networking approach conducted in this study. Chemical family classification is predicted based on the applied dereplication pipeline. Further indicated are total node count and total numbers of different levels of chemical identification determined in this analysis.
Table S12. Phylogenetic and chemical information about the performed metabolic cluster analysis in Myoporeae. Comparison of the chemical content information extracted from determined tanglegram metabolic clusters (TMCs). The underlying metabolic cluster analysis is based on the absence/presence of the 100 largest chemical families of the generated molecular network of Myoporeae. Numbers of specimens from major phylogenetic clades are shown with ‘O’: outgroup, ‘AG’: clades A to G, ‘H3’: clade H3, ‘H10’: clade H10, ‘H11’: clade H11, ‘H12’: clade H12, ‘H14’: clade H14, and ‘H’: remaining clade H‐associated taxa. Dominant chemical families are depicted for each TMC, with ‘CF100’ indicating chemical families present in all species (100%) of the corresponding TMC and ‘CF90’ and ‘CF80’ depicting 90% and 80% abundancy of chemical families among the associated specimens, respectively. Different levels of predicted chemical classification and associated scores are shown. If present, level 1 identifications and their associated molecular structures are displayed.
Table S13. Detailed chemical information about heatmap metabolic clusters. Heatmap metabolic clusters (HMCs) were generated using a categorical heatmap analysis based on the presence/absence of the 100 largest chemical families found in the molecular network of Myoporeae. Information about chemical identity is given based on the conducted dereplication pipeline. Further indicated is spectrometric and structural information about corresponding level 1 identification events.
Table S14. Identified metabolites within the 100 largest chemical families in Myoporeae. Detailed spectrometric and structural information about level 1 and 2 identification events found within the largest 100 chemical families from the molecular network of Myoporeae.
Table S15. Dereplication of serrulatane‐ and viscidane‐related chemical families. A list of 30 chemical families from Myoporeae that have been dereplicated as serrulatane‐ or viscidane‐related. Chemical family classification is predicted based on the applied dereplication pipeline. Further indicated are total node count and total numbers of different levels of chemical identification determined in this analysis as well as mass and retention time median of each chemical family.
Table S16. Predicted serrulatane and viscidane diterpenoid structures in chemical families of Myoporeae. Detailed spectrometric and structural information about serrulatane and viscidane diterpenoid‐related identification events determined within the 30 chemical families within the Myoporeae molecular network that have been found to be associated with this specific chemistry.
Figure S1. Schematic workflow of the chemo‐evolutionary analysis. In total, 291 specimens from species and subspecies of the tribe Myoporeae were investigated regarding their chemical and phylogenetic relationships. LC‐QTOF‐HRMS was applied on crude leaf extracts to generate mass spectrometric data including MS1 and MS2. MZmine2 was used to isolate spectral features from the raw data and align those across all samples to output a feature table that contains the relatively quantified chemical composition of the dataset. Spectral information of those isolated features is then exported and integrated into the feature‐based molecular networking (FBMN) pipeline at GNPS (https://gnps.ucsd.edu). The generated molecular network is composed of nodes and edges that represent individual spectral features and share spectral similarity, respectively. Spectral similar nodes fall into clusters of varying size that are considered as a chemical family of structurally related metabolites. The molecular network provides the foundation for an exhaustive dereplication approach involving public, in‐house, and in silico libraries of reference metabolites for spectral comparison. Dereplicated nodes are structurally annotated and utilized to achieve a prediction of chemical classification for each chemical family by using the Network Annotation Propagation (NAP) method. Using the unsupervised substructure discovery approach MS2LDA, substructural motifs can be predicted within the given spectral data and annotated throughout the molecular network. To compare the chemical profiles of all 291 specimens, the presence (at least one spectral feature present of a corresponding chemical family) and absence (no feature present in the sample) information of chemical families can be used. This approach yields a binary dataset that can be compiled by a metabolic cluster analysis into a dendrogram that displays chemical similarity among the specimens. In contrast, a phylogenetic analysis shows the evolutionary relationships among those specimens and is based on nuclear ribosomal DNA sequencing of leaf tissue from the same specimens. A tanglegram analysis directly compares the analyses by rearranging branches in both dendrograms to reveal similar clustering (i.e., specimens D and E) and thus chemo‐evolutionary relationships. Groups of specimens that display an evolutionary lineage are referred to as phylogenetic clades, while chemical similarity is shared within tanglegram metabolic clusters (TMCs). To extend the view on the chemo‐evolutionary framework, a categorical heatmap analysis is used that clusters the presence/absence of individual chemical families according to their phylogenetic distribution alongside the tanglegram‐refined phylogeny. Heatmap metabolic clusters (HMCs) can be derived from this approach, which represent subsets of chemical families that share a similar phylogenetic signature. Additional functional annotations can be integrated to test metadata for correlation with the chemo‐evolutionary framework that is displayed by the heatmap.
Figure S2. Specimen and peak signal intensity distribution of singleton features. Distribution graphs showing the number of singleton features according to the number of associated specimens and peak signal intensity (detected maximum value for each feature among all specimens). Calculated medians are highlighted by a dotted vertical line and the actual value displayed below each respective graph.
Figure S3. Collection of tanglegram analyses. Conjunction of phylogenetic and metabolic information from 291 species of the tribe Myoporeae visualized by different tanglegram analyses. The nuclear ribosomal DNA‐based phylogeny shows taxa with posterior probability above 95%. Metabolic cluster analyses were conducted based on different datasets: (a) presence/absence of individual spectral features after normalization of the full spectral dataset (10 696 features); (b) presence/absence of the 30 chemical families found to be associated with serrulatane or viscidane diterpenoid metabolism within the generated global molecular network of Myoporeae; (c) peak signal intensity of individual spectral features after normalization of the full spectral dataset (10 696 features); (d) presence/absence of individual spectral features assigned as singletons after normalization; and (e–g) presence/absence of the 100 largest chemical families within the generated molecular network of Myoporeae using an inclusion factor of (e) 2, (f) 3, and (g) 5. Phylogenetic subclades are indicated with the most prevalent ones highlighted by color code. The tanglegram analysis connects the same specimens by a line, which is colored when equal branching of taxa is present in both analyses. Selected species with varying intraspecific chemical variation are highlighted within selected metabolic cluster analyses, with square (E. duttonii), diamond (E. alternifolia), and circle (E. deserti).
Figure S4. Continuous heatmap analysis showing chemo‐evolutionary relationships in Myoporeae. A heatmap analysis was used to put chemical information into an evolutionary context. For that, information about the 100 largest chemical families was compiled, which was derived from the molecular network of Myoporeae. For each specimen in this study, the total amount of metabolites of a corresponding chemical family is displayed within the heatmap. Left: The nuclear ribosomal DNA phylogeny on the left side presents the major phylogenetic clades. Top: As depicted on top of the heatmap, the chemical information underwent hierarchical clustering according to the given phylogeny to reveal chemo‐evolutionary patterns among subsets of chemical families, defined as heatmap metabolic clusters (HMCs) I–X. Bottom: Selected chemical classifications generated by NAP‐based dereplication are displayed below in blue, while the presence of level 1 identification (m/z, retention time, and MS2 match) in a particular chemical family is shown in gold. Right: Functional annotations including the presence of leaf resin and hairiness, pollination, antibacterial activity, traditional medicinal usage, and biogeographical species distribution information are displayed on the right side. A summary of legends is also included to the right of the figure.
Figure S5. Comparison of species distribution between members of TMC A‐ and B‐related chemistry. Species distributions for members of tanglegram metabolite clusters A (A1–A6) and B (B1 and B2). Widespread species (those with broad distributions spanning more than one biome) have been excluded from maps. Species assessed as widespread were identified in clusters A1 (Eremophila bignoniiflora, Eremophila alternifolia, Eremophila deserti), A2 (Myoporum montanum, Eremophila latrobei subsp. glabra, Eremophila deserti), A5 (Myoporum acuminatum, Eremophila longifolia), and B2 (Eremophila mitchellii). Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 29 June 2020.
Figure S6. Geographic distributions of two widespread Eremophila species. Eremophila alternifolia and Eremophila duttonii specimen locations are marked with red boxes. Data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020.
Figure S7. Molecular network analysis of Eremophila deserti. Global molecular network representation of metabolite distribution from three E. deserti specimens. Highlighted in yellow are putative metabolites that are shared by at least two specimen of E. deserti. Metabolites found specifically in one of the specimens are marked in blue: E. deserti (MJB2384), red: E. deserti (RMF228), and green: E. deserti (RMF204). On each node in the network, the total number of specimens that share this specific metabolite is displayed. Indicated on the right side is the geographic distribution of the observed E. deserti specimen in this study, with corresponding sampling locations marked with colored boxes. Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020. Maps of Australian annual mean temperature, annual mean precipitation, and annual mean solar radiation were downloaded as layers for reference from the Atlas of Living Australia (ala.org.au).
Figure S8. Spectral comparison of three Eremophila deserti specimens. Spectral representation of base peak chromatograms for three E. deserti specimens that display interspecific chemical variation. Selected compounds are highlighted and chemical annotations are stated below. An extracted ion chromatogram of specimen RMF204 is also shown to show the complete lack of flavonoid compounds present in specimen MJB2384.
Figure S9. Species distributions for members of Myoporeae phylogenetic clades A–H. In this representation, widespread species (those with broad distributions spanning more than one biome) have been excluded from maps. Species assessed as widespread were identified in phylogenetic clades A (Myoporum acuminatum, Myoporum montanum) D (Eremophila bignoniiflora, Eremophila deserti, Eremophila polyclada), G (Eremophila alternifolia), and H (Eremophila latrobei subsp. glabra, Eremophila longifolia, Eremophila maculata subsp. maculata, Eremophila mitchellii). Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020.
Figure S10. Climatic information about Australia. Maps of Australian annual mean temperature, annual mean precipitation, annual mean solar radiation, and annual mean relative humidity, as well as major classes of the Koppen Climate Classification were downloaded as layers for reference for the species distribution maps in this study from the Atlas of Living Australia (ala.org.au).
Figure S11. Species distributions for members of Myoporeae phylogenetic clade H, subclades H1–H14. In this representation, widespread species (those with broad distributions spanning more than one biome) have been excluded from maps. Species assessed as widespread were identified in phylogenetic subclades H2 (Eremophila mitchellii), H5 (contains only Eremophila longifolia, so no distribution map was included), H8 (Eremophila maculata subsp. maculata), and H10 (Eremophila latrobei subsp. glabra). Species occurrence data were generated from the Australasian Virtual Herbarium (https://avh.chah.org.au/) on 6 April 2020.
Figure S12. Continuous heatmap analysis of chemo‐evolutionary patterns of serrulatane and viscidane diterpenoids in Myoporeae. A heatmap analysis was conducted to put chemical information into an evolutionary context. For that, information about the 30 chemical families derived from the molecular network of Myoporeae which have been found to be associated with serrulatane and viscidane diterpene chemistry was compiled. For each specimen in this study, the total amount of metabolites of a corresponding chemical family is displayed within the heatmap. Left: The nuclear ribosomal DNA phylogeny on the left side presents the major phylogenetic clades. Top: As depicted on top of the heatmap, the chemical information was subjected to hierarchical clustering according to the given phylogeny to reveal chemo‐evolutionary patterns. Bottom: Selected chemical classifications generated by NAP‐based dereplication are displayed below in blue, while the presence of level 1 identification (m/z, retention time, and MS2 match) in a particular chemical family is shown in gold. Right: Functional annotations including the presence of leaf resin and hairiness, pollination, antibacterial activity, traditional medicinal usage, and biogeographical species distribution information, as well as tanglegram metabolic cluster (TMC) associations, are displayed on the right side. A summary of legends is also included to the right of the figure.
Figure S13. Annotation of substructural motif ‘Eremophila motif 440’. Annotation of the unknown substructural motif ‘Eremophila motif 440’ was conducted using the combined information from the biochemical reaction prediction tool MetWork and the in silico MS2 spectrum prediction tool CFM‐ID. (a) The spectral representation of the studied MS2LDA motif indicates the probabilities of presence and absence of distinct fragments, which have been annotated using in silico spectrum prediction by CFM‐ID on serrulatane diterpenoid structures found to be associated with ‘Eremophila motif 440’, such as KU006‐14. Based on these structural annotations a dihydroxy serrulatane acid bicyclic core was proposed as the substructure underlying this motif. (b) MetWork‐based prediction of biochemical and structural reactions joining serrulatane diterpenoid features present in chemical family 10 that are associated with ‘Eremophila motif 440’. The predictions indicate the loss of the motif upon dehydroxylation at the aromatic ring, underlying these specific modifications as the inherent motif characteristics. Nodes highlighted in light green/yellow correspond to prior dereplicated spectral features, while dark green displays unknown features that where structurally annotated by the MetWork analysis.
Figure S14. Spectral representation of MS2LDA‐predicted substructural motifs found in chemical families 10 and 22. Spectral information on the probabilities of mass fragments being present or absent within different substructural motifs was determined in chemical families 10 and 22 by the MS2LDA tool.
Figure S15. Phylogeny of tribe Myoporeae based on analysis of nuclear ribosomal DNA sequences. Bayesian inference 50% majority‐rule consensus tree, showing posterior probability values on each branch. Branches with values of <0.50 collapsed.
Figure S16. LC‐QTOF‐HRMS data quality control. The quality of the full spectral dataset used in this study was assessed by plotting all samples (red) together with the quality controls (blue) within a principal component analysis.
Data Availability Statement
All DNA sequences included in the phylogenetic analysis for this manuscript have been submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with accession numbers given in Table S1. The DNA sequence alignment and resulting tree have been submitted to TreeBase (https://www.treebase.org/treebase‐web/home.html) and are available for reviewer consideration at http://purl.org/phylo/treebase/phylows/study/TB2:S26197?x‐access‐code=8f551ae84c6f32323d73a86cc9e8d1bd&format=html.
Raw spectral data derived from the 291 specimens, additional quality checks, and blank samples are available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000086644.
This MASSIVE database repository also contains the metadata information used in this study (file name ‘Eremophila ChemoEvo metadata complete QC BLANK update.txt’ in the ‘metadata’ tab) as well as the MZmine2 output before (file name ‘20200220 MZMINE OUTPUT.csv’) and after normalization (‘20200220 MZMINE OUTPUT AFTER NORMALISATION.csv’) as well as spectral database files for in silico prediction used during the NAP workflow as supplementary files (‘20200222 Eremophila ISDB.tsv/csv’ within the ‘other’ tab found in ‘Browse Dataset Files’) for download. Metadata information is interpreted as follows: leaf resin: ‘0’ – absence, ‘1’ – presence; leaf hairiness: ‘0’ – absence, ‘1’ – presence; pollination: ‘0’ – insect, ‘1’ – bird; traditional use: ‘0’ – no documented use, ‘1’ – use is documented; antibacterial activity: ‘0’ – tested absence of activity, ‘1’ tested presence of activity (corresponding references are found in Table S1 of the manuscript); biogeographical distribution: ‘0’ – widespread, ‘1’ – Eremean, ‘2’ – Southwestern temperate, ‘3’ – Southeastern temperate, ‘5’ – non‐Australian; general phylogenetic clades: ‘o’ – outgroup, ‘ag’ – clades A to G, ‘h3’ – clade H3, ‘h10’ – clade H10, ‘h11’ – clade H11, ‘h12’ – clade H12, ‘h14’ – clade H14, and ‘h’ – remaining clade H‐associated taxa; sample weight: weight in mg of the plant material used for extraction per sample. Described data files are also found within the digital supplement in addition to all relevant tables from this study in machine‐readable formats as well as compiled in one Excel sheet. In addition, all dendrograms used to generate tanglegrams (Newick format) as well as the underlying data matrices (tab separated) for the metabolic cluster analysis shown in Figure 3 and Figure S3 are provided within the digital supplement. Thereby individual spectral features are often indicated as follows: ‘[compound ID] [mass] [retention time]’.
The molecular networking job can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f74d3374a7ba43eb8d58e5d2a312bf33.
The NAP job can be publicly accessed at https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=5980fea229b9427bb610a8e7d6c25827. The MS2LDA job can be publicly accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8082c8133d1843c39af42164157ab043.
Spectral information derived from the 76 reference compounds used in this study is available at the GNPS spectral library under the CCMSLIB IDs given in Table S3.
Underlying data for geographic distribution maps are available for:
Figure S5 TMC A1: https://doi.ala.org.au/doi/10.26197/ala.337ac334‐016c‐4ad3‐a8dd‐f9ce79aef8a7
TMC A2: https://doi.ala.org.au/doi/10.26197/ala.4693c8c5‐cad1‐4824‐ba5f‐c4d25aa38686
TMC A3: https://doi.ala.org.au/doi/10.26197/ala.a026ad9e‐56a3‐482b‐a2f7‐fa2201b9c3b2
TMC A4: https://doi.ala.org.au/doi/10.26197/ala.edb84e7b‐9e4c‐479e‐b581‐300eb4d27bcc
TMC A5: https://doi.ala.org.au/doi/10.26197/ala.af1cf180‐7c18‐42eb‐93b2‐6dbe8b27d399
TMC A6: https://doi.ala.org.au/doi/10.26197/ala.fdcb84c8‐5db6‐4fea‐8f93‐2101fb0d2af0
TMC B1: https://doi.ala.org.au/doi/10.26197/ala.6ce9b49c‐d67f‐4242‐83ef‐495868db9800
TMC B2: https://doi.ala.org.au/doi/10.26197/ala.2a8e032c‐bbb3‐4bd1‐936a‐2ca83269b1a8
Figure S6 Eremophila alternifolia: https://doi.ala.org.au/doi/10.26197/ala.cacf63e9‐5681‐4581‐af10‐d1c6f4799143
Eremophila duttonii: https://doi.ala.org.au/doi/10.26197/ala.a6acf423‐00b3‐41f4‐8d5c‐dddcc6553f54
Figure S7 Eremophila deserti: https://doi.ala.org.au/doi/10.26197/ala.f2d34675‐c1ae‐4a81‐adf9‐fe03e34cd388
Figure S9 Clade A: https://doi.ala.org.au/doi/10.26197/ala.3e501c4c‐da8547e4‐985f‐2af4d6365766
Clade B: https://doi.ala.org.au/doi/10.26197/ala.3e0044f8‐5383‐4106‐8fb4‐ae0a3a2be0a6
Clade C: https://doi.ala.org.au/doi/10.26197/ala.2528df04‐a91d‐42b8‐88f0‐6f66bfca3f61
Clade D: https://doi.ala.org.au/doi/10.26197/ala.f82d15d5‐f54a‐431b‐b690‐c193553d55c4
Clade E: https://doi.ala.org.au/doi/10.26197/ala.6cc5e4e9‐8560‐4d31‐8550‐6576c0b3b2b0
Clade F: https://doi.ala.org.au/doi/10.26197/ala.99838369‐78e7‐4de5‐aac4‐20feeea4e748
Clade G: https://doi.ala.org.au/doi/10.26197/ala.2d5c0411‐960b‐4d87‐9e98‐186032eb7ccb
Clade H: https://doi.ala.org.au/doi/10.26197/ala.2d2f885c‐ff52‐4b89‐aa00‐b121ab789a66
Figure S11 Clade H1: https://doi.ala.org.au/doi/10.26197/ala.ff62d761‐f339‐4933‐88ae‐0087fdaf84a0
Clade H2: https://doi.ala.org.au/doi/10.26197/ala.e00c3310‐66f5‐42a4‐9df8‐52a5e0cc50ab
Clade H3: https://doi.ala.org.au/doi/10.26197/ala.53afe67f‐cccf‐4dfb‐98cd‐ce3a935bdfb7
Clade H4: https://doi.ala.org.au/doi/10.26197/ala.6a389f01‐9668‐4de5‐af9e‐f83f75478cb8
Clade H6: https://doi.ala.org.au/doi/10.26197/ala.ddf5f8b9‐87a9‐4b77‐b59d‐389ae763231b
Clade H7: https://doi.ala.org.au/doi/10.26197/ala.ef9633ba‐4eef‐43c5‐951b‐f8a065c1033e
Clade H8: https://doi.ala.org.au/doi/10.26197/ala.5fdea1f0‐8854‐4fc0‐b26e‐22cec0d4242c
Clade H9: https://doi.ala.org.au/doi/10.26197/ala.6a655fd9‐a8ee‐4174‐8ffa‐ce7cab21dc12
Clade H10: https://doi.ala.org.au/doi/10.26197/ala.02071a0a‐7523‐4380‐96ac‐9485dc3388db
Clade H11: https://doi.ala.org.au/doi/10.26197/ala.35441d4d‐99c0‐43fa‐a4ee‐eea1bc6160c4
Clade H12: https://doi.ala.org.au/doi/10.26197/ala.c18cf6c9‐eba3‐48ac‐82a5‐13af5983f892
Clade H13: https://doi.ala.org.au/doi/10.26197/ala.6bea9d4f‐7c52‐43c6‐983a‐359e60d01832
Clade H14: https://doi.ala.org.au/doi/10.26197/ala.19793839‐fd96‐4dda‐b552‐d157f1e77af4


