Figure 2.
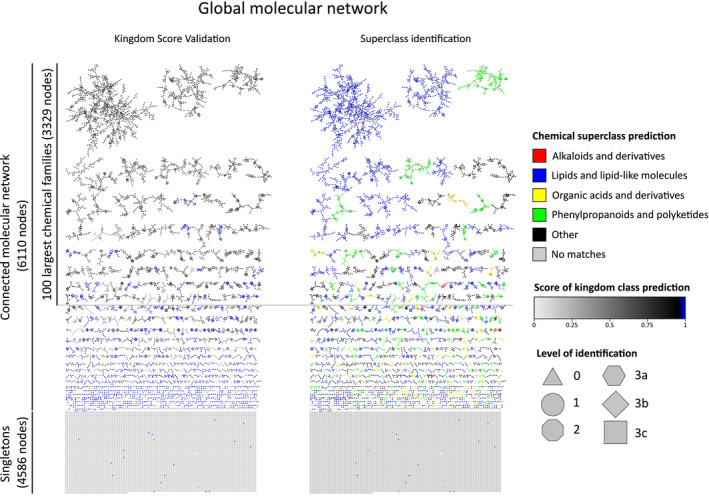
The global molecular network of Myoporeae.
The global molecular network of Myoporeae comprises a connected part, with clusters of spectral features that represent structurally related putative metabolites, as well as singletons, which represent features that did not show spectral similarity to others. The connected dataset was dereplicated using public, in‐house, and in silico spectral libraries and subsequent Network Annotation Propagation (NAP). Identification of nodes took place on six different levels, with level 1: m/z, retention time, and MS2 match using an in‐house spectral library; level 2: MS2 match using in‐house and public spectral libraries; level 3a–c: Fusion, Consensus, or MetFrag algorithm‐based in silico fragmentation database match; level 0: no spectral match. The presented network on the left displays a kingdom score‐based assessment with overall high scoring values that support a robust dereplication of the corresponding chemical families. The network on the right side displays NAP‐generated chemical superclass prediction for each chemical family, with selected classes highlighted in color.
