Summary
Adjustment to energy starvation is crucial to ensure growth and survival. In Arabidopsis thaliana (Arabidopsis), this process relies in part on the phosphorylation of the circadian clock regulator bZIP63 by SUCROSE non‐fermenting RELATED KINASE1 (SnRK1), a key mediator of responses to low energy.
We investigated the effects of mutations in bZIP63 on plant carbon (C) metabolism and growth. Results from phenotypic, transcriptomic and metabolomic analysis of bZIP63 mutants prompted us to investigate the starch accumulation pattern and the expression of genes involved in starch degradation and in the circadian oscillator.
bZIP63 mutation impairs growth under light‐dark cycles, but not under constant light. The reduced growth likely results from the accentuated C depletion towards the end of the night, which is caused by the accelerated starch degradation of bZIP63 mutants. The diel expression pattern of bZIP63 is dictated by both the circadian clock and energy levels, which could determine the changes in the circadian expression of clock and starch metabolic genes observed in bZIP63 mutants.
We conclude that bZIP63 composes a regulatory interface between the metabolic and circadian control of starch breakdown to optimize C usage and plant growth.
Keywords: Arabidopsis, bZIP63, circadian clock, growth, low energy stress, starch
Introduction
Plants rely on a sophisticated network of metabolic and environmental receptors that trigger downstream signaling pathways to cope efficiently with environmental changes and ensure survival. Efficient management of nutrient and energy resources (i.e. mainly carbohydrates but also amino acids and fatty acids, all of which can fuel respiration to produce ATP) is crucial to accommodate growth, development and stress responses, counting on the adequate regulation of anabolic and catabolic processes (Baena‐González et al., 2007; Baena‐González & Sheen, 2008; Robaglia et al., 2012; Lastdrager et al., 2014; Tomé et al., 2014). The balance between anabolism and catabolism is regulated mostly by the antagonistic activity of two evolutionary conserved kinases, namely SUCROSE non‐fermenting RELATED KINASE1 (SnRK1) and TARGET OF RAPAMYCIN (TOR), which repress and activate growth under conditions of low and high energy levels, respectively (Baena‐González et al., 2007; Robaglia et al., 2012; Dobrenel et al., 2016). The usage of photosynthetically produced carbohydrates in Arabidopsis is tightly controlled to maintain a steady supply of energy (Gibon et al., 2004; Graf et al., 2010; Stitt & Zeeman, 2012). Photosynthates are partitioned into sucrose and transitory starch, which is mostly consumed during the dark period to sustain metabolic activities and growth (Lu et al., 2005; Smith & Stitt, 2007; Graf et al., 2010; Kölling et al., 2015).
In Arabidopsis, dynamic adjustment of the starch degradation requires a functional circadian clock to ensure proper supply of carbohydrates until dawn (Gibon et al., 2004; Baena‐González et al., 2007; Graf et al., 2010; Scialdone et al., 2013; Seki et al., 2017), but the molecular players and the mechanisms involved in this regulation are mostly unknown. The plant circadian clock provides a way to predict daily and seasonal environmental changes associated with the movements of the Earth (i.e. rhythmic changes in light availability and temperature), allowing optimization of metabolic activities, development, growth and responses to stress (Green et al., 2002; Dodd et al., 2005; Harmer, 2009; Seo & Mas, 2015; Shim & Imaizumi, 2015). Mathematical models have been proposed to account for the empirical data of starch turnover dynamics regulated by the circadian clock. To describe the mechanisms by which the appropriate rate of starch degradation is set by the circadian clock, some models focus on the detection of the starch amount (Scialdone et al., 2013; Pokhilko et al., 2014; Scialdone & Howard, 2015), whereas others focus on the perception and maintenance of sucrose homeostasis (Haydon et al., 2013; Seki et al., 2017).
We have shown that the Arabidopsis transcription factor bZIP63, which is a key target of SnRK1, regulates some of the transcriptional changes induced by energy deprivation (Baena‐González et al., 2007; Matiolli et al., 2011; Mair et al., 2015) and also mediates the circadian clock entrainment by sugars through regulation of PSEUDO‐RESPONSE REGULATOR 7 (PRR7) transcription (Frank et al., 2018). Here we present evidence that both the circadian clock and the energy status regulate bZIP63 transcript accumulation. This dual regulation of bZIP63 expression may, in turn, affect its target genes expression, including those involved in starch degradation and energy deficit responses. Accordingly, bZIP63 mutants displayed alteration in starch degradation and impaired growth in diel cycles. Our results suggest that bZIP63 is a link between energy status and the circadian clock to control starch degradation and responses to energy stress, fine‐tuning carbon utilization through the diel cycle.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana Wassilewskija (Ws) and Columbia‐0 (Col‐0) ecotypes, as well as the T‐DNA insertion mutants bzip63‐2 (FLAG_610A08; Matiolli et al., 2011; Supporting Information Fig. S1a–d) and bzip63‐3 (FLAG_532A10; Fig. S1a,b), were obtained from the Arabidopsis Biological Resource Center (ABRC). The mutants cca1‐11/lhy‐21 (cca1/lhy; ABRC germplasm CS9380, Ws background; Graf et al., 2010) (CCA1, CIRCADIAN CLOCK ASSOCIATED 1; LHY, Late Elongated Hypocotyl), phosphoglucomutase (pgm) (Col‐0 background; Gibon et al., 2004), and the overexpressing lines HA‐bZIP63‐ox1 and HA‐bZIP63‐ox2 (Frank et al., 2018) were described previously. In all experiments, plants were grown in a 2:1 mix of substrate (Plantmax HT, São Paulo, Brazil) and fine vermiculite (Plantmax). Sown seeds were kept in the dark at 4°C for 72 h to break dormancy, and afterwards were grown at 22°C and Photosynthetically Active Radiation (PAR) of 100 μmol m−2 s−1 under short day (SD, 10 h : 14 h, light : dark), long day (LD, 16 h : 8 h, light : dark), equinoctial (12 h : 12 h, light : dark) or free‐running (LL) photoperiods.
For transcriptome analysis and validation of misregulated genes by quantitative reverse transcription PCR (qRT‐PCR) analysis, total RNA was extracted from the four youngest leaves and the shoot apical meristems of plants grown under SD conditions for 25 d, and harvested at the end of the night (EN) immediately before the onset of the light, because bZIP63 is most expressed in meristem and young leaves (Weltmeier et al., 2009) and at EN (DIURNAL Database: http://diurnal.mocklerlab.org; Fig. S1e). For chromatin immunoprecipitation (ChIP) analysis, 1 g of the entire aerial part of plants grown under equinoctial conditions for 12 d were harvested at EN. For gene expression analyses under diel and free‐running conditions, plants were grown under equinoctial conditions for 30 d. Then, samples from the entire aerial part were harvested every 4 h for 2 d, whereas another set of plants was released into free‐running conditions for 24 h before being sampled every 4 h for 48 h (24–72 h, LL) and snap‐frozen in liquid N2 for subsequent total RNA extraction. For metabolic profiling analysis, plants were grown under equinoctial conditions for 25 d and entire aerial parts were harvested at the end of the night immediately before the onset of the light. Each biological replicate was composed of a pool sampled from five plants for gene expression analysis or six plants for metabolic profiling analysis. Phenotypic analyses were performed in 30‐d‐old plants grown under SD, LD, equinoctial or LL conditions, and in Phenoscope platform (https://phenoscope.versailles.inra.fr; Tisné et al., 2013) plants grown under 8 h : 16 h, light : dark photoperiod.
RNA isolation, cDNA synthesis and qRT‐PCR analysis
Total RNA for the transcriptome analysis was isolated using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions. The purity of total RNA samples was verified by spectrophotometry (260/230 and 260/280 ratios ≥ 1.8; NanoVue, GE Healthcare Life Sciences, Piscataway, NJ, USA). RNA integrity was evaluated by capillary electrophoresis (RNA integrity number > 8, Bioanalyzer 2100; Agilent, Santa Clara, CA, USA). Total RNA isolation for qRT‐PCR was performed as described previously (Oñate‐Sánchez & Vicente‐Carbajosa, 2008), with modifications (Oliveira et al., 2015). For gene expression quantification of intron‐less genes, RNA was treated with Ambion® Turbo DNA‐free DNAse (cat. no. AM1907; Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. cDNA synthesis from 1.5 µg total RNA (final volume 12.5 ml) was performed using ImProm II Reverse Transcriptase (cat. no. A3802; Promega) and oligo(dT)18 according to the manufacturer’s instructions. qRT‐PCR analyses were performed as described previously (Matiolli et al., 2011), using Platinum SYBR green (cat. no. 11733‐038; Invitrogen), and run on a 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). PP2AA3 (AT1G13320) or ACTIN2 (AT3G18780) were used as reference genes as indicated in the text (Czechowski et al., 2005).
Microarray and data analysis
Three biological replicates of each genotype (Wassilewskija (Ws) and bzip63‐2) were used for transcriptome analysis. Each replicate was composed of a pool of the four youngest leaves and shoot apical meristem harvested from five plants. Hybridization on the GeneChip Arabidopsis ATH1 array was done in the LNBio (Brazilian Biosciences National Laboratory, Campinas, Brazil) Microarray Facility (LMA). Robust Multi‐array Average (RMA) normalization was performed using Expression Console™ (Affymetrix) and statistical data analysis was performed using the affylmGUI R package (R Core Team, 2018). A P‐value < 0.05 was used as the cutoff for the selection of differentially expressed genes and expression level between genotypes. Overlaps with previously available transcriptome data were carried out using the webtool Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/), and statistical significance of overlaps were estimated using a web‐based software designed by Jim Lund (University of Kentucky), with statistical significance quantified using a hypergeometric test (Kim et al., 2001; http://nemates.org/MA/progs/overlap_stats.cgi). The heatmap in Fig. 1(f) was obtained using multiexperiment viewer (Saeed et al., 2003) and over‐representation of biological pathways were analyzed using mapman (Thimm et al., 2004). Microarray data are available at GEO (accession number GSE119175).
Fig. 1.
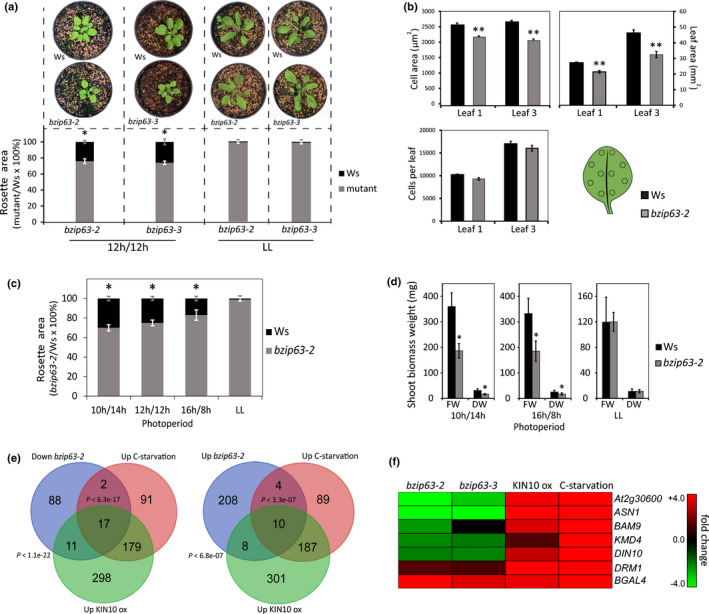
Arabidopsis bZIP63 mutants present reduced growth under diel cycles. (a) The mutants bzip63‐2 and bzip63‐3 exhibit reduced rosette area when grown under diel cycles (12 h : 12 h, light : dark photoperiod). The rosette area of bZIP63 mutants and Wassilewskija (Ws) were indistinguishable when plants were grown under free‐running conditions (LL). (b) Leaf epidermal cell properties of bzip63‐2 compared to Ws grown under diel cycles (12 h : 12 h, light : dark photoperiod). Leaf area was measured directly whereas the number of cells in the leaf epidermis and epidermal cell area were calculated from a series of measurements corresponding to c. 10% of the total leaf area taken from 10 different positions across the leaf using an Eclipse 80i light microscope, and displayed as the mean of 10 samples obtained from two independent experiments with the corresponding SE values. Leaf order was counted as the oldest leaf developed after the cotyledons onwards (two‐tailed Student’s t‐test; n = 20; **, P < 0.01). (c) Differences in rosette area between bzip63‐2 and Ws are inversely correlated to photoperiod length, where short days (SD; 10 h : 14 h, light : dark photoperiod) unveil the most contrasting rosette areas between bzip63‐2 and Ws. The data were obtained from three and two independent experiments for bzip63‐2 and bzip63‐3, respectively, with 30 biological replicates for each genotype (two‐tailed Student’s t‐test; n = 90 for bzip63‐2 and 60 for bzip63‐3; *, P < 0.05). Values are the percentage of mean rosette area in bZIP63 mutants compared to Ws, and error bars indicate standard deviation. (d) FW and DW differences between bzip63‐2 and Ws are inversely correlated to photoperiod length, where SDs promoted the most contrasting shoot biomass weight. Under LL no difference for both FW and DW was observed between genotypes. Plants were grown for 30 d under photoperiods or 25 d under LL. The biomass weight was obtained from two independent experiments, with 30 biological replicates each (Student’s t‐test; n = 60; *, P < 0.05). Values are means, and error bars indicate standard deviation. (e) Overlap between genome‐wide misregulated genes in bzip63‐2, genes induced by KIN10 overexpression (KIN10 ox; Baena‐González et al., 2007) and various carbon (C) starvation conditions (Contento et al., 2004; Gibon et al., 2004; Usadel et al., 2008; Cookson et al., 2016). (f) Clustering of energy stress associated genes misregulated in bzip63‐2, induced by KIN10 ox and by various C starvation conditions and selected for validation in bzip63‐3 through quantitative reverse transcription PCR.
ChIP
Samples were vacuum‐infiltrated with 1% formaldehyde solution in water and incubated at room temperature for 20 min to allow the crosslink of DNA‐protein complexes. After cross‐linking, samples were ground in liquid N2 using mortar and pestle. Extraction of DNA–protein complexes was performed as described previously (Gendrel et al., 2002) and resuspended in 300 μl nuclear lysis buffer (50 mM Tris‐HCL, pH 8; 10 mM EDTA; 1% SDS; 1× Pierce protease inhibitor cocktail #88265 (cat. no. A32963; Thermo Fisher Scientific). Re‐suspended DNA‐protein complexes were sonicated in a Bioruptor® sonication system (Diagenode, Denville, NJ, USA) using the program, nine cycles of 30 min ON/1 min OFF with high potency, to achieve DNA fragments ranging from 75 to 500 bp, which was verified by agarose gel electrophoresis. DNA‐protein complexes were isolated using EpiQuik™ Plant ChIP kit (cat. no. P‐2014‐48; Epigentek Group Inc., Farmingdale, NY, USA) following the manufacturer’s instructions. Monoclonal anti‐HA antibody (cat. no. sc‐7392 C1313; Santa Cruz Biotechnology, TX, USA) combined with the Plant ChIP kit were used to capture HA‐tagged bZIP63‐DNA complexes. qPCR target enrichment analyses were performed using Platinum SYBR green (cat. no. 11733‐038; Invitrogen) run on a 7500 Applied Biosystems Fast Real‐time PCR System. Two transgenic lines overexpressing HA‐tagged bZIP63, HA‐bZIP63‐ox1 and HA‐bZIP63‐ox2 (Fig. 2f; Frank et al., 2018), were used for ChIP‐qPCR experiments. Overexpressor lines have been used previously for ChIP (e.g. Portolés & Más, 2010; Zheng et al., 2015; Frank et al., 2018); the two overexpressor lines accumulate approximately three‐fold more bZIP63 transcript than the wild‐type (WT) at EN (Frank et al., 2018). Data were normalized using cycle threshold (Ct) derived from the comparison of the Ct of the anti‐HA antibody IP samples (adjusted relative to Ct of input DNA) and the Ct of mock IP (adjusted relative to Ct of the input DNA); Student’s t‐tests compared between the control and the anti‐HA treated samples as described previously (Haring et al., 2007).
Fig. 2.
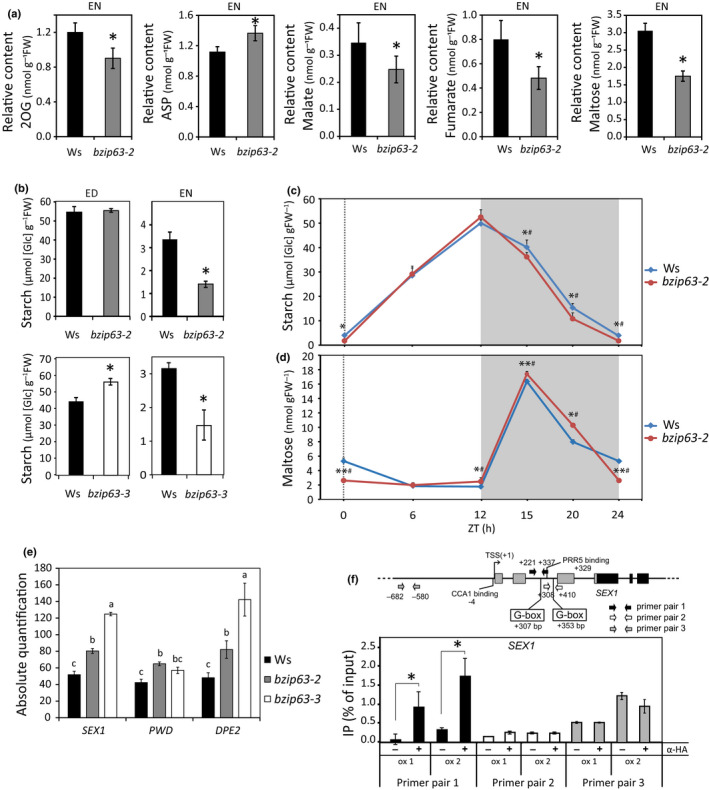
bZIP63 modulates starch degradation. (a) Relative metabolite content at the end of the night (EN; ZT = 24 h) in leaves of bzip63‐2 as compared to Wassilewskija (Ws). The concentrations of amino acids, organic acids and sugar were determined by gas chromatography time‐of‐flight mass spectroscopy (GC‐TOF‐MS). The average amount of metabolites was measured with five biological replicates, and each replicate consisted of leaves collected from six 25‐d‐old plants (two‐tailed Student’s t‐test; n = 30; *, P < 0.05). (b) Starch amounts at the end of the day (ED; ZT = 12 h) and EN (ZT = 24 h) in leaves of bzip63‐2 and bzip63‐3 mutants as compared to Ws. The average amount of starch for each time point were measured from two independent experiments, with five biological replicates, and each replicate consisted of leaves collected from six 30‐d‐old plants (two‐tailed Student’s t‐test; n = 60; *, P < 0.05; **, P < 0.01). Time‐course analysis of (c) starch and (d) maltose amounts determined by high‐performance liquid chromatography (HPLC) in bzip63‐2 and Ws leaves through the diel cycle (12 h : 12 h, light : dark photoperiod). The average amount of starch and maltose for each time point were measured from two independent experiments, with five biological replicates, and each replicate consisted of leaves collected from six 30‐d‐old plants (two‐tailed Student’s t‐test; n = 60 for starch and 30 for maltose; *, P < 0.05; **, P < 0.01; and Wilcoxon’s test #, P < 0.05). (e) Absolute transcript levels of starch degradation‐related genes STARCH EXCESS1 (SEX1), PHOSPHOGLUCAN, WATER DIKINASE (PWD) and DISPROPORTIONATING ENZYME 2 (DPE2), are higher in bzip63‐2 and bzip63‐3 at the end of the night (EN). Data were obtained from three independent experiments, with three biological replicates, and each replicate consisted of four youngest leaves and the shoot apical meristems collected from five 25‐d‐old plants, differences were tested performing ANOVA and the different letters indicate groups that are significantly different (Tukey’s honestly significant difference (HSD) test, P < 0.05). (f) Enrichment of specific regions of SEX1 5´sequences by chromation immunoprecipitation (ChIP) in HA‐bZIP63‐ox1 and HA‐bZIP63‐ox2 transgenic lines. bZIP63 binding to SEX1 cis regulatory sequences was assessed by ChIP using anti‐HA antibody followed by quantitative PCR. Scheme of SEX1 gene structure, depicting the binding sites of the circadian clock transcriptional regulators CCA1 and PRR5, as well as the bZIP63 binding regions containing canonical G‐box motifs. Primer pair 1 amplifies a 5′ region containing the closer G‐box motif relative to the transcription start site (TSS+1), whereas primer pair 2 amplifies a region containing the second closer G‐box motif relative to TSS+1, and the primer pair 3 amplifies a region without G‐box (−) indicates mock and (+) indicates immunoprecipitated samples. Data were obtained from four independent experiments for HA‐bZIP63‐ox1 and seven for HA‐bZIP63‐ox2 (Student’s t‐test; *, P < 0.05) for primer pairs 1 and 2, and one experiment for primer pair 3. Values are means, and error bars indicate standard deviation.
Phenotypic analysis
The leaf and rosette area (cm2) were measured using Leaf Area Meter 3100 (Li‐Cor Inc., Lincoln, NE, USA) or Phenoscope platform. For FW and DW measurements, rosettes were harvested, and the FW measured immediately. Afterwards, the same samples were dried in an oven at 70°C, and weighing was performed every 24 h until the samples reached a constant DW. Thirty plants were used for each genotype, discarding the three upper and lower outliers for each genotype. The epidermal cell area and the number of the rosette leaves were performed in an Eclipse 80i light microscope (Nikon, Tokyo, Japan). Samples were fixed using methanol, then cleared using lactic acid and mounted for analysis. The number of epidermal cells was obtained by counting the cells inside an area taken from 10 different positions across the leaf, corresponding to c. 10% of the total leaf area (leaves 1 and 3), and the total leaf area were measured, and from these values, the number of cells in the leaf epidermis was extrapolated. The Kolmogorov–Smirnov test was performed to ensure that the data were normally distributed and then Student’s t‐test was performed to identify significant differences between the bzip63‐2 and Ws (P < 0.01).
Extraction and quantification of starch
Starch was determined in the insoluble material after ethanolic extraction of soluble sugars, followed by enzymatic digestion with α‐amyloglucosidase and α‐amylase (Hendriks et al., 2003). Whole rosettes from six 30‐d‐old plants were used as a single sample. Samples were harvested immediately frozen in liquid N2, ground to a fine powder, and 20 mg FW measured and kept at −80°C until analysis.
Metabolite profiling analysis
Forty milligrams of the grounded plant tissue were used for MTBE : methanol : water 3 : 1 : 1 (v/v/v) extraction, as described previously (Giavalisco et al., 2011). The 150 μl of the organic phase was dried and derivatized according to (Roessner et al., 2001). Then 1 μl derivatized samples were analyzed on a Combi‐PAL autosampler (Agilent Technologies) coupled to an Agilent 7890 gas chromatograph (GC) coupled to a Leco Pegasus 2 time‐of‐flight mass spectrometer (TOF‐MS) (LECO, St Joseph, MI, USA). Chromatograms were exported from Leco chromaTOF software (v.3.25) to R software. Peak detection, retention time alignment, and library matching were performed using R/targetsearch (Cuadros‐Inostroza et al., 2009). Metabolites were quantified by the peak intensity of a selective mass. Metabolite intensities were normalized by the FW, followed by the sum of total ion count. Each metabolite value was further normalized by the median of this given metabolite in all measured samples. For maltose, in addition to quantification by the method described above, another method based on high‐pressure anion‐exchange liquid chromatography (HPLC) separation was used as described in (Martins et al., 2013; Fig. 2d). The whole rosettes from six 30‐d‐old plants were used as a single sample. Samples were harvested and immediately frozen in liquid N2, and 100 mg grounded tissue was kept at −80°C until analysis.
Statistical analyses
All statistical tests, n number, the measure of the means and the error bars are described in figure legends when appropriate. For comparison between the two groups, two‐tailed Student’s t‐test and Wilcoxon’s test were used. For multiple comparison statistical tests ANOVA followed by the Tukey’s honestly significant difference (HSD) test were performed using genes (Cruz, 2013). A hypergeometric test (Kim et al., 2001) was used to estimate the statistical significance of the overlap between transcriptomes. Analyses were considered significant at: *, P < 0.05; **, P < 0.01. Circadian oscillation parameters were calculated from two 24 h cycles under LL, excluding the first 24 h of data. The period was estimated using COSOPT and JTK_CYCLE, the significance thresholds were set to an adjusted P < 0.05 for JTK_CYCLE and a pMMC‐β < 0.05 for COSOPT (Hughes et al., 2010; Yang & Su, 2010). The phase and amplitude were estimated using meta cycle 2D with P < 0.05.
Results
bZIP63 mutants display a photoperiod‐dependent growth impairment
bZIP63 mediates energy stress responses triggered by SnRK1 (Baena‐González et al., 2007; Mair et al., 2015; Dröge‐Laser & Weiste, 2018), which suggests that bZIP63 has an important role in energy management. Thus, we hypothesized that disruption of bZIP63 expression could affect plant growth, especially when environmental conditions restrain photosynthesis or respiration, therefore limiting carbohydrate and ATP production. To verify this hypothesis, we performed a comparative analysis of the growth and development of bZIP63 mutants and their respective WT, the Ws ecotype. We found that 30‐d‐old plants of the T‐DNA insertion mutants bzip63‐2 and bzip63‐3 had a rosette area nearly 25% smaller than Ws when grown under equinoctial conditions, but not under constant free‐running conditions (i.e. LL; Fig. 1a), and that the reduction of rosette size in the mutant was correlated with smaller leaf and epidermal cell area (Fig. 1b).
Because bZIP63 is involved in the adjustment to an energy deficit, we reasoned that changes in photoperiod, and hence C availability through the day (Lu et al., 2005; Haydon et al., 2013; Dröge‐Laser & Weiste, 2018), could have an impact on the growth of bZIP63 mutants. Indeed, the most contrasting rosette size shown by bzip63‐2 and Ws was observed on plants grown in SD conditions, where bzip63‐2 leaf and rosette area and DW were reduced by 30% and 46%, respectively (Figs 1c,d, S2b; Fig. S2a shows growth in 8 h : 16 h, light : dark photoperiod). In LD conditions, bzip63‐2 leaf and rosette area and DW were reduced by only 17% and 30%, respectively, compared to Ws (Figs 1c,d, S2b). When plants were grown under LL conditions, the rosette area and weight of bzip63‐2, bzip63‐3 and Ws were indistinguishable (Figs 1a,c,d). Altogether, the phenotypic data suggest that changes in photoperiod and the resulting alterations of C supply affect the impact of bZIP63 on plant growth performance.
bZIP63 binds to energy deficit‐responsive genes
In order to obtain clues about the underlying reason of the growth impairment in the bZIP63 mutant, we performed a comparative gene expression analysis of young leaves of bzip63‐2 and Ws grown under SD and harvested at EN (= ZT 24), immediately before the onset of the light, to maximize the discovery of genes misregulated in bzip63‐2 (Fig. S1e). The resulting gene expression profiles revealed 230 upregulated and 118 downregulated genes in bzip63‐2 (Table S1). Among the downregulated genes in bzip63‐2, there was a 23% (P < 1.1e‐22) overlap with the genes induced by KIN10 overexpression, the SnRK1 catalytic subunit (Baena‐González et al., 2007; Table S1; Fig. 1e), which is consistent with the role of bZIP63 in mediating KIN10‐induced transcriptional changes (Baena‐González et al., 2007; Mair et al., 2015). Noticeably, ASPARAGINE SYNTHASE 1 (ASN1), DARK INDUCIBLE 10 (DIN10), BETA‐AMYLASE 9 (BAM9), At2g30600 and KISS ME DEADLY 4 (KMD4/At3g59940), that are induced by KIN10 and by C starvation (Contento et al., 2004; Gibon et al., 2004; Baena‐González et al., 2007; Usadel et al., 2008; Cookson et al., 2016), and can therefore be considered as low C/energy marker genes, were repressed in bzip63‐2 (Fig. 1f). ASN1, BAM9 and At2g30600 were induced in two transgenic lines overexpressing HA:VP16:bZIP63 fusion (HA‐bZIP63‐ox1 and HA‐bZIP63‐ox2; Table S2). Target enrichment analysis using ChIP followed by qPCR showed that bZIP63 binds to the 5′‐sequences of these genes in vivo (Fig. S3a–c), suggesting that they are direct targets of bZIP63. These results corroborate the role of bZIP63 as a key transcription factor acting downstream of KIN10 to reprogram transcription in response to low energy stress (Baena‐González et al., 2007; Matiolli et al., 2011; Mair et al., 2015).
bZIP63 mutants show faster starch degradation during the night
A subset of 10 genes that are induced by KIN10 overexpression (P < 6.8e‐07; Baena‐González et al., 2007) and various conditions of C depletion (P < 3.3e‐07; Contento et al., 2004; Gibon et al., 2004; Usadel et al., 2008; Cookson et al., 2016; Fig. 1e), and therefore also can be considered as low C/energy marker genes, were induced in bzip63‐2 (Fig. 1e,f). This result suggests that the management of energy supply is impaired in bzip63‐2. To further evaluate this possibility, we performed a comparative primary metabolic profiling analysis between bzip63‐2 and WT at EN using GC‐TOF‐MS (Table S3). We found that bzip63‐2 had a reduction of the concentrations of 2‐oxoglutarate (2OG) (Fig. 2a), an intermediate of the TCA cycle and an important C‐skeleton for glutamine synthesis by glutamate synthase from glutamate in N assimilation. 2OG therefore is considered to integrate C and N metabolism (Nunes‐Nesi et al., 2010). The reduced 2OG content by contrast with the unchanged concentrations of glutamate and glutamine is consistent with the limiting C availability (Nunes‐Nesi et al., 2010; Fig. 2a; Table S3). Furthermore, larger amounts of aspartate (ASP) in the mutant than in the WT (Fig. 2a), also may reflect a decrease in available sugars at the EN (Nunes‐Nesi et al., 2010). Additionally, malate and fumarate, which can fuel respiration to provide energy when starch reserves are exhausted (Zell et al., 2010), also were decreased in bzip63‐2 (Fig. 2a), further suggesting a superimposed shortage of C/energy. Finally, maltose, the major product of starch breakdown, was diminished in the mutant (Fig. 2a), suggesting faster exhaustion of starch reserves. Indeed, bzip63‐2 and bzip63‐3 had 50% less starch at EN compared to Ws (Fig. 2b). The amount of starch accumulated in these mutants was similar (bzip63‐2, Fig. 2b) or slightly higher (bzip63‐3, Fig. 2b) at the end of the day (ED). These results suggest that bzip63‐2 and bzip63‐3 degrade starch faster during the night. To get a more detailed picture of how starch metabolism is affected by bZIP63, we performed a time‐course analysis of starch and maltose, measured by HPLC, in bzip63‐2 during a 24 h diel cycle (equinoctial conditions, Fig. 2c,d; SD, Fig. S4a,b). The differences in starch content between bzip63‐2 and Ws were significant as early as 3 h after the beginning of the night (ZT15, Fig. 2c; ZT13, Fig. S4a), suggesting that bZIP63 mutation affects starch degradation since ED. The amounts of maltose in bzip63‐2 were higher than those observed for Ws at the beginning of the night (i.e. ZT15 and ZT20; Fig. 2d), which correlates with the faster starch breakdown found in this mutant compared to Ws (Fig. 2c). The lower concentrations of maltose in bzip63‐2 at EN measured by both HPLC and GC‐TOF‐MS most likely results from a reduction of its production as a consequence of premature exhaustion of starch reserves (Fig. 2a,d).
We found that the transcript levels of two starch phosphorylating enzymes, namely α‐GLUCAN, WATER DIKINASE1/STARCH EXCESS1 (GWD1/SEX1) and PHOSPHOGLUCAN, WATER DIKINASE (PWD), which are required for starch degradation (Mahlow et al., 2014), were higher in both bzip63‐2 and bzip63‐3 mutants at EN (Figs 2e, S4c; Tables S1, S2, S4). In addition, the cytosolic maltotriose‐metabolizing enzyme DISPROPORTIONATING ENZYME 2 (DPE2), an essential component of the pathway that releases glucose from maltose (an intermediate of starch breakdown; Fettke et al., 2006), also was induced in both bzip63‐2 and bzip63‐3 (Figs 2e, S4c; Tables S1,S2). Subsequent ChIP‐qPCR analysis showed that VP16:HA‐tagged bZIP63 binds to the 5′ sequence of SEX1 (Fig. 2f), suggesting that this gene is a direct target of bZIP63. These data establish a positive correlation between higher SEX1, PWD and DPE2 transcript levels and faster starch degradation in the mutants. Altogether, the results indicate that the faster starch degradation in the bZIP63 mutants and the consequent early depletion of starch at the EN (Figs 2b,c, S4) led to low energy stress, which may explain – at least in part – the growth defect of these mutants. This hypothesis is consistent with the notion that C/energy starvation towards dawn impairs growth (Smith & Stitt, 2007; Graf et al., 2010; Apelt et al., 2017).
The circadian clock and C/energy status interact to regulate bZIP63 diel expression pattern
The circadian clock is crucial to define the rate of starch breakdown compatible with a regular C supply during the diel cycle (Lu et al., 2005; Graf et al., 2010; Scialdone et al., 2013; Pokhilko et al., 2014; Seki et al., 2017; Seaton et al., 2018). We demonstrated that bZIP63 is essential for circadian clock entrainment by sugars, transducing this metabolic signal into the core oscillator through the transcriptional regulation of the clock component PSEUDO‐RESPONSE REGULATOR (PRR)7 (Frank et al., 2018). In addition, previous reports showed that the circadian clock transcriptional repressors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), PRR5 and PRR7 bind to the bZIP63 promoter (Nagel et al., 2015; Liu et al., 2016), and that bZIP63 expression is higher in both the prr579 triple mutant (Liu et al., 2016) and the cca1/lhy double mutant (Graf et al., 2017) when compared to the respective WT accessions, suggesting that bZIP63 expression is under circadian clock control. Thus, we investigated in more detail the reciprocal regulation between bZIP63 and the circadian clock, aiming to shed light on how starch degradation and energy stress responses are regulated in a timely manner. We first verified whether bZIP63 expression is regulated by the circadian clock. Indeed, bZIP63 transcript level oscillates in free‐running conditions in both Ws and the circadian clock mutant cca1/lhy (Graf et al., 2010), closely matching the Ws period (c. 24 h) and the shorter period (c. 17 h) of cca1/lhy (Fig. 3a,g; Table S5). As expected, the expression of the circadian clock‐regulated gene GRANULE BOUND STARCH SYNTHASE 1 (GBS1; Graf et al., 2010) showed a shorter oscillation period in free‐running conditions and a phase advance in diel conditions in the cca1/lhy mutant as compared to Ws (Fig. 3b,g; Table S5). These results demonstrate that bZIP63 is under circadian clock control. The similarity of bZIP63 and BAM9 expression profiles (Fig. 3a,c) is compatible with BAM9 being a target of bZIP63 (Figs 1f, S3; Table S2).
Fig. 3.
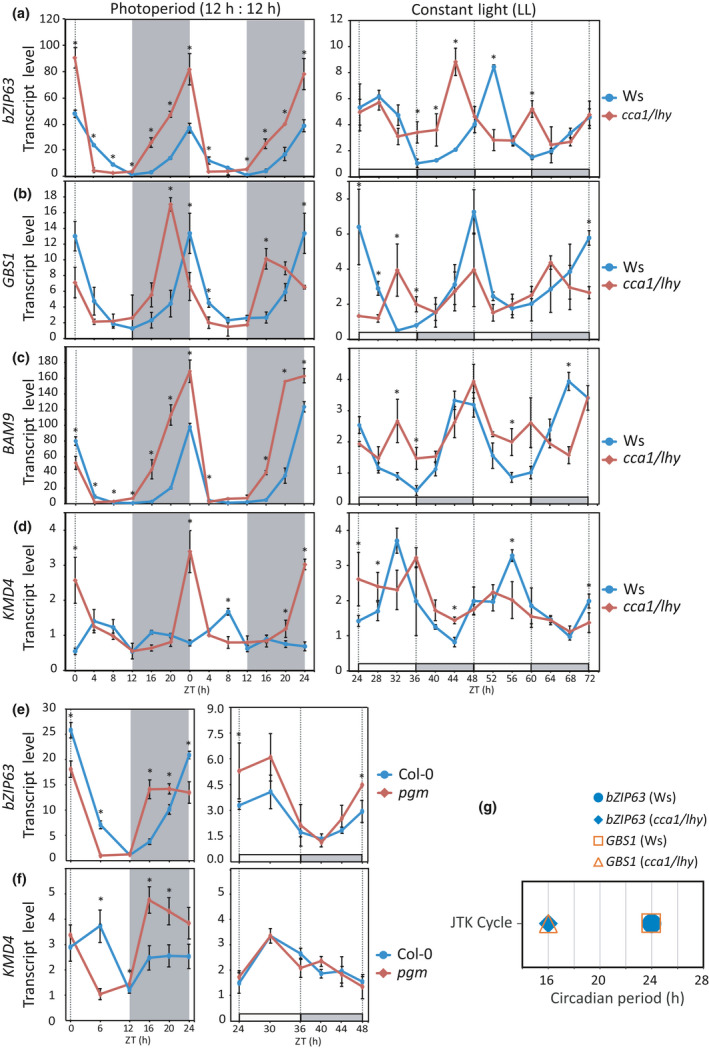
The circadian clock and carbon (C)/energy status control bZIP63 oscillatory pattern. (a) Circadian oscillation of bZIP63 transcript levels follows the shorter period of the double mutant cca1/lhy under free‐running conditions (LL), but does not show the expected phase advance under photoperiod (CCA1, CIRCADIAN CLOCK ASSOCIATED 1; LHY, Late Elongated Hypocotyl). (b) Transcripts levels of GBS1, which is a gene under circadian clock regulation, follows the shorter period of cca1/lhy in LL and shows a phase advance in photoperiod. (c) The bZIP63 direct target BAM9 presents transcript level oscillation following the shorter period of cca1/lhy in LL, but does not have the phase advance under photoperiod. (d) Under photoperiod, the C/energy starvation marker KMD4 is induced during the last part of the night in cca1/lhy. (e) bZIP63 transcript oscillation pattern is altered in the starch synthesis deficient mutant pgm under photoperiod, showing a rapid increase in the first hours of the night, which is consistent with the decrease of sugar concentrations at this time (Gibon et al., 2004). By contrast, transcript levels of bZIP63 were similar between pgm and Col‐0 in the early subjective night in LL. (f) The transcript level of the starvation marker KMD4 is increased at early night in pgm, whereas this is not observed in the early subjective night in LL. (g) Transcript levels of bZIP63 and GBS1 follow the shorter period of the double mutant cca1/lhy under LL. In the plotted graphs of photoperiod, white or gray backgrounds represents light or dark period, respectively. In graphs of LL, the x‐axis represents the time elapsed after releasing the plants into LL. The subjective day (ZT 24–36, 48–60) and subjective night (ZT 36–48, 60–72) are identified by white and gray bars on the x‐axis, respectively. The data were obtained from two independent experiments, with three biological replicates at each time point, and each replicate consisted of the whole rosette collected from three plants (two‐tailed Student’s t‐test; n = 9; *, P < 0.05). Values are means, and error bars indicate standard deviation.
Interestingly, in the cca1/lhy mutant grown in diel conditions, bZIP63 transcript level – by contrast to that of GBS1 – peaked at dawn as in Ws (Fig. 3a,b). Moreover, the bZIP63 transcript level oscillation showed a higher amplitude in cca1/lhy than in Ws (ZT16; Fig. 3a). We reasoned that, as bZIP63 expression is regulated by C/energy levels (Baena‐González et al., 2007; Matiolli et al., 2011; Mair et al., 2015), its oscillation pattern in cca1/lhy grown in light : dark cycles could be due to C/energy starvation towards EN as a consequence of the premature exhaustion of starch reserves (Graf et al., 2010). Energy starvation in cca1/lhy was supported by the induction of the C/energy starvation marker gene KMD4 (Graf et al., 2010) in the last hours of the night under photoperiodic growth conditions (Fig. 3d). In addition, the phase advance of bZIP63 expression in the starchless mutant pgm under photoperiod (Fig. 3e) reinforces the notion that bZIP63 expression is regulated by C/energy levels and the circadian clock. The pgm mutant does not accumulate any significant amount of starch and, therefore, experiences reduced C and energy availability early in the night (Gibon et al., 2004), as demonstrated by the stronger induction of KMD4 in this mutant (Fig. 3f). Under LL conditions, in which C availability is not limiting, no differences in bZIP63 and KMD4 expression were found between Col‐0 and pgm (Fig. 3e,f). These results provide strong support for the notion that bZIP63 transcript oscillation pattern (i.e. phase and amplitude) is set by the energy levels and the circadian clock.
Interaction between bZIP63 and circadian clock regulates starch degradation‐related genes and energy deficit‐responsive genes
We observed that 60% of the misregulated genes in bzip63‐2 mutant oscillate under LL conditions (Table S1), which represents a two‐fold enrichment compared to circadian clock‐controlled oscillating genes in Arabidopsis (Covington et al., 2008; Harmer, 2009). These findings suggest that the interaction between bZIP63 and the circadian clock (Fig. 3; Frank et al., 2018) impacts the bZIP63 transcriptional output. Thus, we investigated the importance of this interaction on the expression pattern of genes related to starch degradation and energy deficit responses (Tables S1, S2).
We showed that the transcript levels of the starch degradation‐related genes SEX1, PWD and DPE2 are regulated by bZIP63, as they are induced in bzip63‐2 and bzip63‐3 mutants at EN (Fig. 2e; Table S2). Because the expression of these genes also is known to be regulated by the circadian clock (Ni et al., 2009; Edwards et al., 2010; Seo et al., 2012), we would expect their circadian oscillation pattern to be altered in bZIP63 mutants. Thus, we performed a comparative analysis of SEX1, PWD and DPE2 transcript level oscillation in bzip63‐2 and Ws leaves from plants released into LL conditions rather than under SD or LD cycles, to circumvent possible confounding effects related to energy fluctuations in diel conditions (Fig. 4a; Table S2).
Fig. 4.
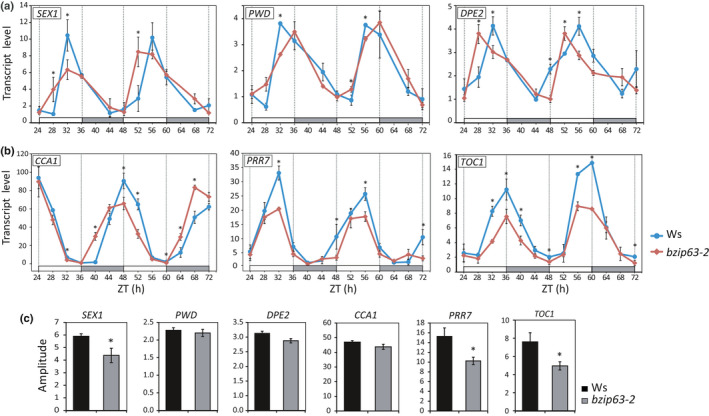
bZIP63 modulates the oscillatory pattern of circadian clock and starch degradation related genes. Transcript oscillatory pattern of starch degradation related genes (a), and circadian clock genes (b), are altered in bzip63‐2 as compared to Ws. 30‐d‐old plants were entrained under 12 h : 12 h, light : dark photoperiod and released into free‐running conditions (LL) for 3 d. The x‐axis represents the time elapsed after release of the plants into LL, where subjective day (ZT 24–36; 48–60) and subjective night (ZT 36–48; 60–72) are identified by white and gray bars on the x‐axis, respectively. Transcript oscillation amplitude of starch degradation‐related genes STARCH EXCESS1 (SEX1), PHOSPHOGLUCAN, WATER DIKINASE (PWD) and DISPROPORTIONATING ENZYME 2 (DPE2) and the circadian clock genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), PSEUDO‐RESPONSE REGULATOR 7 (PRR7) and TIMING OF CAB EXPRESSION 1 (TOC1) under LL (c). The data were obtained from two independent experiments, with three biological replicates in each time point, and each replicate consisted of the whole rosette collected from three plants (two‐tailed Student’s t‐test; n = 9; *, P < 0.05). Values are means, and error bars indicate standard deviation.
All three genes exhibited different oscillation patterns in bzip63‐2 compared to Ws (Fig. 4a), with lower oscillation amplitude for SEX1 (Fig. 4c; Table S5), and phase advance and delay for DPE2 and PWD transcripts, respectively, in bzip63‐2 (Fig. 4a; Table S5). No significant differences in SEX1 and PWD mRNA levels between WT and bzip63‐2 were detected at the subjective EN under LL conditions (Fig. 4a) as would be expected from the data under diel growth conditions (Fig. 2e). This discrepancy could be related to differences of unknown regulatory features between diel and free‐running growth conditions, such as those deriving from changes in the pattern of C flow.
Expression of SEX1 is deregulated in bZIP63 mutants, and bZIP63 binds to SEX1 5′‐ sequences (Fig. 2e,f). Moreover, the circadian clock component PRR5 was shown to bind next to the region bounded by bZIP63 in SEX1 5′‐ sequences (Liu et al., 2016; Fig. 2f). In addition, CCA1 also was found to bind to the SEX1 promoter (Nagel et al., 2015; Fig. 2f). Therefore, regulation of SEX1 expression could be partly driven by direct targeting of bZIP63 and components of the circadian oscillator to its 5´sequences (Fig. 2f). Because we were unable to show that bZIP63 binds to PWD and DPE2 promoters, it is possible that bZIP63 indirectly regulated the expression of these two genes by influencing the circadian oscillator through regulation of PRR7 expression (Frank et al., 2018; Fig. 4b,c). Indeed, under LL conditions, we found that the phase of CCA1 transcript oscillation was advanced in the bzip63‐2 mutant (Fig. 4b), which is consistent with downregulation of its transcriptional repressor PRR7 in bzip63‐2 (Frank et al., 2018; Fig. 4b,c; Table S2). PRR7 and TIMING OF CAB EXPRESSION 1 (TOC1), which are involved, respectively, in the morning and central loops of the core clock oscillator, oscillate with lower amplitude in bzip63‐2 (Fig. 4b,c). All of these changes indicate that bZIP63 could modulate the circadian expression of PWD and DPE2 through the adjustment of the circadian clock. Therefore, these results suggest that bZIP63 activity can affect the circadian expression of SEX1, PWD and DPE2 by both direct (e.g. by binding to SEX1 5′‐sequences) and indirect mechanisms by adjusting the circadian oscillator and – consequently – its gene expression output.
We found that 28 downregulated genes in bzip63‐2 (23%, P < 1.1e‐22) overlapped with genes induced by KIN10 overexpression (i.e. energy stress responsive genes, Fig. 1e; Table S1). Among these genes, 11 (41%, P < 1.1e‐19) oscillate with the same phase as bZIP63 (ZT 23) under entrainment conditions (Mockler et al., 2007; Fig. S5). This observation raises the possibility that, in a similar way to the starch degradation‐related genes, the expression of a subset of KIN10‐induced energy‐stress responsive genes is controlled by the interaction between bZIP63 and the circadian clock. This assumption was supported by the observation that the upstream regions of two of these KIN10‐induced energy‐stress responsive genes, namely BAM9 (Figs 1f, 3c) and At2g30600 (Fig. 1f) are bound by bZIP63 (Fig. S3b,c) in a region overlapping the sequences bound by both PRR5 and PRR7, and PRR5, respectively (Liu et al., 2016). This dual regulatory scheme of bZIP63 and the circadian clock could explain the higher amplitude of the rhythmic oscillation of BAM9 transcript levels in cca1/lhy under photoperiod (Fig. 3c) probably as a consequence of energy limitation resulting from premature exhaustion of starch (Graf et al., 2010). Altogether, the evidence suggests that bZIP63 misregulation has a broad impact on the circadian oscillation of core oscillator genes, which modulates the circadian clock output, such as starch metabolism and responses to low energy signals.
Discussion
During daylight, plants harvest energy from sunlight to synthesize soluble sugars to support metabolism and growth. A fraction of the photoassimilates is saved in the form of storage carbohydrates, such as the leaf transitory starch, to ensure energy supply during the night. Appropriate management of energy resources is crucial to promote growth and ensure reproduction, thus increasing fitness. We show here that the transcription factor bZIP63, a key phosphorylation target of SUCROSE non‐fermenting RELATED KINASE1 (SnRK1) (Baena‐González et al., 2007; Mair et al., 2015), is essential for optimal growth, probably through its involvement in managing energy resources. By comparison with the wild‐type (WT), bzip63‐2 and bzip63‐3 mutants exhibited faster night‐time starch degradation, which was correlated with lower amounts of metabolites related to carbon (C) status (Fig. 2a; Table S3), indicating a situation of energy deficit at the end of the night (EN). Reduction of available energy in bZIP63 mutants at EN also is supported by the induction of the expression of C/energy‐starvation related genes such as Beta‐Galactosidase 4 and Dormancy‐associated protein 1 (Fig. 1e,f; Table S2). Our data are in agreement with previous results showing that even a small reduction in the starch content after an artificial extension of the night period resulted in a significant reduction of carbohydrates and organic acids and an induction of energy deficit marker genes (Gibon et al., 2004; Usadel et al., 2008; Graf et al., 2010). Thus, in a similar way to other mutants impaired in starch metabolism, such as phosphoglucomutase (pgm), STARCH EXCESS1 (sex1‐1), BETA‐AMYLASE 3 (bam3) and bam4 (Fulton et al., 2008; Paparelli et al., 2013), or the circadian clock mutant cca1/lhy (Graf et al., 2010) (CCA1, Circadian Clock Associated 1; LHY, Late Elongated Hypocotyl), the reduced growth phenotype observed in bzip63‐2 and bzip63‐3 possibly is due to reduced C supply, and the resulting energy deficit, towards EN (Graf et al., 2010; Kölling et al., 2015; Mair et al., 2015). This conclusion is supported by the equalization of WT and bZIP63 mutants growth under constant light, which provides a stable photosynthate supply throughout the day. A similar phenomenon has been described for the starch metabolism mutants pgm and sex1‐1 (Izumi et al., 2013). The involvement of bZIP63 in the growth optimization seems to be more critical under short day (SD) conditions (Fig. 1c,d), where less energy is available in a 24 h period (Sulpice et al., 2014), partly because less starch is accumulated at the end of the day (ED) in SD conditions in comparison to long day (LD) conditions (Fig. S6a). In addition, the small amount of starch at EN of SDs in bzip63‐2 (Fig. S6a) most likely resulted in a stronger energy starvation, which is supported by the greater accumulation of transcripts of the energy deficit marker ASN1 (Fig. S6b). Thus, accentuated growth impairment of bzip63‐2 in SD conditions (Figs 1c,d, S2a,b) is likely a consequence of a stronger energy deficit towards the EN. These results indicate that bZIP63 is involved in the regulation of starch degradation, which supports the proposed role of bZIP63 in C and energy metabolism management (Baena‐González et al., 2007; Mair et al., 2015). The reduced bzip63‐2 mutant leaf cell size (Fig. 1b) can partly explain the smaller leaf area of this mutant, and is correlated with the downregulation of cell wall remodeling XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE genes (Table S1). One of them, XTH15, whose expression was correlated with cell elongation (Pedmale et al., 2016), is regulated by sugar concentrations (Matiolli et al., 2011) and was found to be involved in cell wall remodeling (Hayashi & Kaida, 2011). XTH15 also was found to be regulated by bZIP63 (Fig. S7a,b), and also by the circadian oscillator through the circadian clock component PRR5 (Liu et al., 2016). Hence, these results suggest that bZIP63 may directly participate in the regulation of cell expansion.
Starch degradation is controlled by the circadian clock to maintain a constant sugar supply required for growth during the night (Smith & Stitt, 2007; Graf et al., 2010; Sulpice et al., 2014; Flis et al., 2016). The faster starch degradation observed in bzip63‐2 and bzip63‐3 is correlated with the upregulation of genes encoding the starch degradation‐related enzymes PHOSPHOGLUCAN, WATER DIKINASE (PWD) and DISPROPORTIONATING ENZYME 2 (DPE2) at EN but a direct causal relationship remains to be established. In any case, the diel fluctuation of SEX1, PWD and DPE2 transcript levels possibly is regulated by the circadian clock through direct binding of CCA1 and PRR5 to their promoters (Nagel et al., 2015; Liu et al., 2016), and bZIP63 because the phase or amplitude of the oscillation pattern of these transcripts were changed in bzip63‐2 under free‐running (LL) conditions (Fig. 4a,c; Table S5). bZIP63 was shown to have a direct input on the circadian clock by participating in its entrainment by sugars (Frank et al., 2018). In turn, as shown here, the circadian clock feeds back to regulate bZIP63 expression, which combined with C/energy levels, shapes the diel oscillation pattern of bZIP63 transcript levels (Fig. 3a,e). Thus, bZIP63 is part of a regulatory module that integrates C/energy and circadian clock signals to fine‐tune the circadian transcript level oscillation of the starch degradation‐related genes PWD, SEX1 and DPE2. This regulation may be achieved by the direct regulation of target genes, as suggested by bZIP63 binding to a SEX1 5′ sequence containing a canonical bZIP G‐box binding site localized next to a PSEUDO‐RESPONSE REGULATOR 5 (PRR5) target region (Fig. 2f). Alternatively, bZIP63 may act indirectly, such as in the case of PWD and DPE2, through the regulation of the circadian oscillator in response to sugars (Frank et al., 2018; Figs 2e, 4a,c). The same regulatory rationale may also apply to 22 of 28 (78%) C/energy deficit responsive genes downregulated in bzip63‐2 (i.e. induced by KIN10 overexpression; Baena‐González et al., 2007; Fig. 1e; Table S1), which were shown to be bound by at least one of the core oscillator genes CCA1, PRR5, PRR7, PRR9 and TIMING OF CAB EXPRESSION 1 (TOC1) (Nagel et al., 2015; Liu et al., 2016; Table S6). Indeed, we show that bZIP63 binding regions co‐localize with sequences bound by circadian clock components in the 5′ sequences of the energy stress marker genes BAM9 and At2g30600 (Edwards et al., 2010; Liu et al., 2016; Fig. S3b,c; Table S6). Based on these observations, we speculate that bZIP63, interacting with PRRs, is involved in defining the oscillation pattern of downstream genes as part of a transcriptional gating mechanism similar to the co‐occupancy of circadian clock‐regulated genes by PRRs and the PHYTOCHROME INTERACTING FACTORS PIF3 and PIF4 to mediate the photosensory pathway and thermoresponsive growth, respectively (Soy et al., 2016; Zhu et al., 2016; Martín et al., 2018).
bZIP63 also may have opposite regulatory activities because the expression of SEX1 and stress‐related genes (e.g. BAM9) are up‐ and downregulated, respectively, in bZIP63 mutants (Fig. 1e, 2e). This antagonistic regulatory feature may depend on the architecture of the promoter that bZIP63 binds to and consequently its interaction with other transcription factors. For instance, the circadian clock regulator CCA1‐HIKING EXPEDITION (CHE), one of the protein interactors of bZIP63 (Frank et al., 2018), can bind to the promoters of ISOCHORISMATE SYNTHASE 1 (ICS1) and CCA1, acting as an activator and repressor, respectively (Pruneda‐Paz et al., 2009; Zheng et al., 2015). bZIP63 regulatory output also may be modulated by its interaction with a set of other transcription factors (i.e. TCP2, 4, 10 and 14, NAC066, AtMYB56, AtWOX13 and type B Response Regulators ARR18; Veerabagu et al., 2014; Trigg et al., 2017). Finally, bZIP63‐related transcriptional network also will be shaped by its pattern of heterodimerization with the S‐group bZIP1, 2 or 53, which to some extent involves its phosphorylation by SnRK1 (Ehlert et al., 2006; Mair et al., 2015; Pedrotti et al., 2018).
We provide evidence that bZIP63 interacts with the circadian clock to establish the rate of starch degradation, which will ensure adequate C/energy supply throughout the diel cycle and, therefore, optimize growth performance. We suggest that the integration of C/energy levels and circadian clock‐related signals by bZIP63 makes it a player in acclimation responses to environmental‐induced changes in C/energy availability (Fig. 5).
Fig. 5.
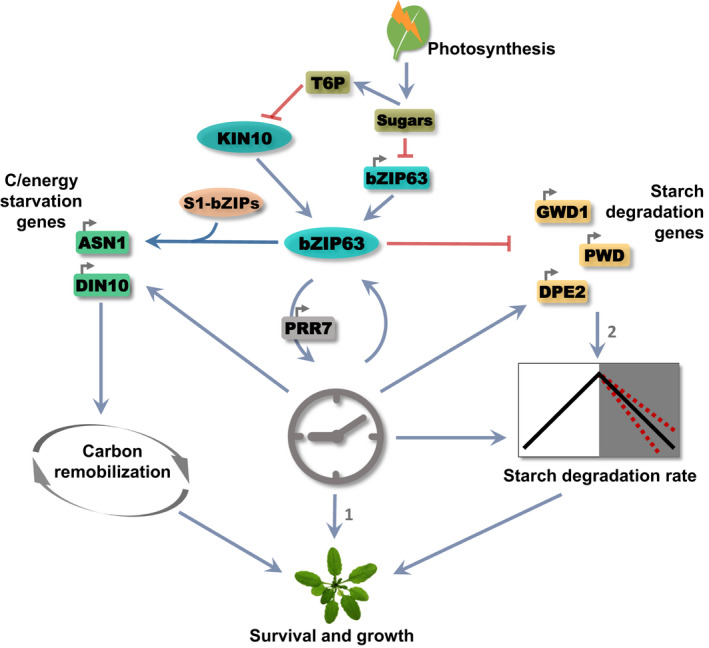
bZIP63 regulates plant growth. The circadian clock and sugars deriving from photosynthesis set the bZIP63 expression pattern. The regulation of energy starvation and starch degradation‐related genes, and the circadian clock gene PSEUDO‐RESPONSE REGULATOR 7 (PRR7) expression by bZIP63 may depend upon its phosphorylation by KIN10, which induces the bZIP63/S1‐bZIPs network (Mair et al., 2015). KIN10 activity is inhibited by Trehalose‐6‐Phosphate (T6P), a proxy of sucrose concentrations (Frank et al., 2018; Zhai et al., 2018). The model proposes two possible modes by which bZIP63 modulates growth: through carbon remobilization to cope with stress or by regulating PRR7 expression to adjust the pace of the circadian clock in response to sugars which would affect the starch degradation rate. These processes are likely to be interlocked by the daily sugar and circadian clock dynamics: (1) circadian clock impacts growth (Dodd et al., 2005; Nozue et al., 2007; Farré, 2012); (2) the direct impact of STARCH EXCESS1 (SEX1), PHOSPHOGLUCAN, WATER DIKINASE (PWD) and DISPROPORTIONATING ENZYME 2 (DPE2) induction in bZIP63 mutants on the starch degradation rate is unknown. Blunt ended arrows indicate repression while arrows indicate activation.
Author contributions
AJCV and CCM designed experiments, collected and analyzed all of the data; DWN performed ChIP‐qPCR and analyzed the data; MCMM and CC performed and analyzed metabolomics data and advised the interpretation; CTH analyzed transcript oscillation data and advised the interpretation; GTD and JGPV performed an experiment under diel condition; EG performed and analyzed the phenotypic experiment on Phenoscope platform; MV designed experiments, analyzed all of the data and obtained the funding; and AJCV, CCM, CC and MV wrote the manuscript. AJCV and CCM contributed equally to this work.
Supporting information
Fig. S1 Characterization of bZIP63 mutants.
Fig. S2 bzip63‐2 mutant has a reduced leaf area and growth rate.
Fig. S3 bZIP63 binds to energy stress‐responsive genes.
Fig. S4 bzip63‐2 starch degradation pattern under SD conditions.
Fig. S5 Circadian phase of KIN10‐induced energy‐stress responsive genes.
Fig. S6 Starch availability in SD vs 12 h : 12 h photoperiods.
Fig. S7 bZIP63 binds to cell wall modification gene.
Table S1 Up‐ and downregulated genes in bzip63‐2 plants compared with the WT.
Table S2 Genes related to circadian clock, starch degradation and energy stress deregulated in bZIP63 mutants.
Table S3 Relative metabolite content at EN in leaves of bzip63‐2 as compared to Ws.
Table S4 List of primers used for quantification of the mRNA levels and the immunoprecipitated sequences by qRT‐PCR.
Table S5 Circadian parameters calculated from two 24‐h cycles under LL conditions.
Table S6 Circadian oscillator binding in downregulated genes in bzip63‐2 that are induced by KIN10 overexpression.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was supported by the São Paulo Research Foundation (FAPESP) (grant nos. 2008/52071‐0, 2012/09351‐8, 2014/04117‐2 and 2015/06260‐0; BIOEN Program), 2018/25710‐4 (FAPESP/UKRI‐BBSRC cooperation agreements), 2019/25993‐9 (FAPESP/Max Planck Institute cooperation agreements) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grants 313104/2017‐4 and 405520/2016‐6).
References
- Apelt F, Breuer D, Olas JJ, Annunziata MG, Flis A, Nikoloski Z, Kragler F, Stitt M. 2017. Circadian, carbon, and light control of expansion growth and leaf movement. Plant Physiology 174: 1949–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena‐González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Baena‐González E, Sheen J. 2008. Convergent energy and stress signaling. Trends in Plant Science 13: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Kim S‐J, Bassham DC. 2004. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiology 135: 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Yadav UP, Klie S, Morcuende R, Usadel B, Lunn JE, Stitt M. 2016. Temporal kinetics of the transcriptional response to carbon depletion and sucrose readdition in Arabidopsis seedlings. Plant, Cell & Environment 39: 768–786. [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. 2008. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biology 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz CD. 2013. GENES – Software para análise de dados em estatística experimental e em genética quantitativa. Acta Scientiarum – Agronomy 35: 271–276. [Google Scholar]
- Cuadros‐Inostroza Á, Caldana C, Redestig H, Kusano M, Lisec J, Peña‐Cortés H, Willmitzer L, Hannah MA. 2009. TargetSearch – a Bioconductor package for the efficient preprocessing of GC‐MS metabolite profiling data. BMC Bioinformatics 10: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W‐R. 2005. Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C. 2016. TOR signaling and nutrient sensing. Annual Review of Plant Biology 67: 261–285. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. 2005. Cell biology: plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Dröge‐Laser W, Weiste C. 2018. The C/S1 bZIP network: A regulatory hub orchestrating plant energy homeostasis. Trends in Plant Science 23: 422–433. [DOI] [PubMed] [Google Scholar]
- Edwards KD, Akman OE, Knox K, Lumsden PJ, Thomson AW, Brown PE, Pokhilko A, Kozma‐Bognar L, Nagy F, Rand DA et al. 2010. Quantitative analysis of regulatory flexibility under changing environmental conditions. Molecular Systems Biology 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente‐Carbajosa J, Dröge‐Laser W. 2006. Two‐hybrid protein‐protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. The Plant Journal 46: 890–900. [DOI] [PubMed] [Google Scholar]
- Farré EM. 2012. The regulation of plant growth by the circadian clock. Plant Biology 14: 401–410. [DOI] [PubMed] [Google Scholar]
- Fettke J, Chia T, Eckermann N, Smith A, Steup M. 2006. A transglucosidase necessary for starch degradation and maltose metabolism in leaves at night acts on cytosolic heteroglycans (SHG). The Plant Journal 46: 668–684. [DOI] [PubMed] [Google Scholar]
- Flis A, Sulpice R, Seaton DD, Ivakov AA, Liput M, Abel C, Millar AJ, Stitt M. 2016. Photoperiod‐dependent changes in the phase of core clock transcripts and global transcriptional outputs at dawn and dusk in Arabidopsis. Plant, Cell & Environment 39: 1955–1981. [DOI] [PubMed] [Google Scholar]
- Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE, Wells Newman D, Yochikawa A, Cano‐Ramirez DL, Chembath A et al. 2018. Circadian entrainment in Arabidopsis by the sugar‐responsive transcription factor bZIP63. Current Biology 28: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G et al. 2008. β‐AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β‐amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. 2002. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1 . Science 297: 1871–1873. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L. 2011. Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high‐resolution mass spectrometry. The Plant Journal 68: 364–376. [DOI] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios‐Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M. 2004. Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post‐translational activation of ADP‐glucose pyrophosphorylase in the followin. The Plant Journal 39: 847–862. [DOI] [PubMed] [Google Scholar]
- Graf A, Coman D, Uhrig RG, Walsh S, Flis A, Stitt M, Gruissem W. 2017. Parallel analysis of Arabidopsis circadian clock mutants reveals different scales of transcriptome and proteome regulation. Open Biology 7: 160333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. 2010. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proceedings of the National Academy of Sciences, USA 107: 9458–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM. 2002. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiology 129: 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. 2007. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. 2009. The circadian system in higher plants. Annual Review of Plant Biology 60: 357–377. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Kaida R. 2011. Functions of xyloglucan in plant cells. Molecular Plant 4: 17–24. [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AAR. 2013. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P. 2003. ADP‐glucose pyrophosphorylase is activated by posttranslational redox‐modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiology 133: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. 2010. JTK‐CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome‐scale data sets. Journal of Biological Rhythms 25: 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Makino A, Ishida H. 2013. Autophagy contributes to nighttime energy availability for growth in Arabidopsis . Plant Physiology 161: 1682–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. 2001. A gene expression map for Caenorhabditis elegans . Science 293: 2087–2092. [DOI] [PubMed] [Google Scholar]
- Kölling K, Thalmann M, Müller A, Jenny C, Zeeman SC. 2015. Carbon partitioning in Arabidopsis thaliana is a dynamic process controlled by the plants metabolic status and its circadian clock. Plant, Cell & Environment 38: 1965–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastdrager J, Hanson J, Smeekens S. 2014. Sugar signals and the control of plant growth and development. Journal of Experimental Botany 65: 799–807. [DOI] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu M‐JJ, Shiu S‐HH, Farre EM, Farré EM. 2016. A G‐box‐like motif is necessary for transcriptional regulation by circadian pseudo‐response regulators in Arabidopsis . Plant Physiology 170: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. 2005. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiology 138: 2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlow S, Hejazi M, Kuhnert F, Garz A, Brust H, Baumann O, Fettke J. 2014. Phosphorylation of transitory starch by α‐glucan, water dikinase during starch turnover affects the surface properties and morphology of starch granules. New Phytologist 203: 495–507. [DOI] [PubMed] [Google Scholar]
- Mair A, Pedrotti L, Wurzinger B, Anrather D, Simeunovic A, Weiste C, Valerio C, Dietrich K, Kirchler T, Nägele T et al. 2015. SnRK1‐triggered switch of bZIP63 dimerization mediates the low‐energy response in plants. eLife 4: 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Rovira A, Veciana N, Soy J, Toledo‐Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadí D et al. 2018. Circadian waves of transcriptional repression shape PIF‐regulated photoperiod‐responsive growth in Arabidopsis . Current Biology 28: 311–318. [DOI] [PubMed] [Google Scholar]
- Martins MCM, Hejazi M, Fettke J, Steup M, Feil R, Krause U, Arrivault S, Vosloh D, Figueroa CM, Ivakov A et al. 2013. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6‐phosphate. Plant Physiology 163: 1142–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiolli CC, Tomaz JP, Duarte GT, Prado FM, Del Bem LEV, Silveira AB, Gauer L, Corrêa LGG, Drumond RD, Viana AJC et al. 2011. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiology 157: 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD. 2007. The Diurnal project : diurnal and circadian expression profiling, model‐based pattern matching, and promoter analysis. CSH Symposia LXXII 353–363. [DOI] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda‐Paz JL, Schmitz RJ, Ecker JR, Kay SA. 2015. Genome‐wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis . Proceedings of the National Academy of Sciences, USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. 2007. Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Nunes‐Nesi A, Fernie AR, Stitt M. 2010. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Molecular Plant 3: 973–996. [DOI] [PubMed] [Google Scholar]
- Oliveira RR, Viana AJC, Reátegui ACE, Vincentz MGA. 2015. An efficient method for simultaneous extraction of high‐quality RNA and DNA from various plant tissues. Genetics and Molecular Research 14: 18828–18838. [DOI] [PubMed] [Google Scholar]
- Oñate‐Sánchez L, Vicente‐Carbajosa J. 2008. DNA‐free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparelli E, Parlanti S, Gonzali S, Novi G, Mariotti L, Ceccarelli N, van Dongen JT , Kölling K, Zeeman SC, Perata P et al. 2013. Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis . Plant Cell 25: 3760–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale U, Huang S‐S, Zander M, Cole B, Hetzel J, Ljung K, Reis P, Sridevi P, Nito K, Nery J et al. 2016. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti L, Weiste C, Nägele T, Wolf E, Lorenzin F, Dietrich K, Mair A, Weckwerth W, Teige M, Baena‐González E et al. 2018. Snf1‐RELATED KINASE1‐controlled C/S1‐bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 30: 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Flis A, Sulpice R, Stitt M, Ebenhöh O. 2014. Adjustment of carbon fluxes to light conditions regulates the daily turnover of starch in plants: A computational model. Molecular BioSystems 10: 613–627. [DOI] [PubMed] [Google Scholar]
- Portolés S, Más P. 2010. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis . PLoS Genetics 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda‐Paz JL, Breton G, Para A, Kay SA. 2009. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. 2012. Sensing nutrient and energy status by SnRK1 and TOR kinases. Current Opinion in Plant Biology 15: 301–307. [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR. 2001. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M et al. 2003. TM4: a free, open‐source system for microarray data management and analysis. BioTechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Scialdone A, Howard M. 2015. How plants manage food reserves at night: quantitative models and open questions. Frontiers in Plant Science 6: doi: 10.3389/fpls.2015.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A, Mugford ST, Feike D, Skeffngton A, Borrill P, Graf A, Smith AM, Howard M. 2013. Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife 2: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton DD, Graf A, Baerenfaller K, Stitt M, Millar AJ, Gruissem W. 2018. Photoperiodic control of the Arabidopsis proteome reveals a translational coincidence mechanism. Molecular Systems Biology 14: e7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ohara T, Hearn TJ, Frank A, Da Silva VCH, Caldana C, Webb AAR, Satake A. 2017. Adjustment of the Arabidopsis circadian oscillator by sugar signalling dictates the regulation of starch metabolism. Scientific Reports 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Mas P. 2015. Stressing the role of the plant circadian clock. Trends in Plant Science 20: 230–237. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park M‐J, Lim M‐H, Kim S‐G, Lee M, Baldwin IT, Park C‐M. 2012. A self‐regulatory circuit of CIRCADIAN CLOCK‐ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis . Plant Cell 24: 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Imaizumi T. 2015. Circadian clock and photoperiodic response in Arabidopsis: From seasonal flowering to redox homeostasis. Biochemistry 54: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M. 2007. Coordination of carbon supply and plant growth. Plant, Cell & Environment 30: 1126–1149. [DOI] [PubMed] [Google Scholar]
- Soy J, Leivar P, González‐Schain N, Martín G, Diaz C, Sentandreu M, Al‐Sady B, Quail PH, Monte E. 2016. Molecular convergence of clock and photosensory pathways through PIF3‐TOC1 interaction and co‐occupancy of target promoters. Proceedings of the National Academy of Sciences, USA 113: 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC. 2012. Starch turnover: Pathways, regulation and role in growth. Current Opinion in Plant Biology 15: 282–292. [DOI] [PubMed] [Google Scholar]
- Sulpice R, Flis A, Ivakov AA, Apelt F, Krohn N, Encke B, Abel C, Feil R, Lunn JE, Stitt M. 2014. Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Molecular Plant 7: 137–155. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Tisné S, Serrand Y, Bach L, Gilbault E, Ben Ameur R, Balasse H, Voisin R, Bouchez D, Durand‐Tardif M, Guerche P et al. 2013. Phenoscope: An automated large‐scale phenotyping platform offering high spatial homogeneity. The Plant Journal 74: 534–544. [DOI] [PubMed] [Google Scholar]
- Tomé F, Nägele T, Adamo M, Garg A, Marco‐llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A et al. 2014. The low energy signaling network. Frontiers in Plant Science 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg SA, Garza RM, Macwilliams A, Nery JR, Bartlett A, Castanon R, Goubil A, Feeney J, Malley RO, Huang SC, et al. 2017. CrY2H‐seq : a massively multiplexed assay for deep‐coverage interactome mapping. Nature Methods 14, 819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. 2008. Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiology 146: 1834–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerabagu M, Kirchler T, Elgass K, Stadelhofer B, Stahl M, Harter K, Mira‐Rodado V, Chaban C. 2014a. The interaction of the arabidopsis response regulator ARR18 with bZIP63 mediates the regulation of PROLINE DEHYDROGENASE expression. Molecular Plant 7: 1560–1577. [DOI] [PubMed] [Google Scholar]
- Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schütze K, Wang X, Chaban C, Hanson J, Teige M, Harter K et al. 2009. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Molecular Biology 69: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Su Z. 2010. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 26: 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell MB, Fahnenstich H, Maier A, Saigo M, Voznesenskaya EV, Edwards GE, Andreo C, Schleifenbaum F, Zell C, Drincovich MF et al. 2010. Analysis of Arabidopsis with highly reduced levels of malate and fumarate sheds light on the role of these organic acids as storage carbon molecules. Plant Physiology 152: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J. 2018. Trehalose 6‐phosphate positively regulates fatty acid synthesis by stabilizing wrinkled. Plant Cell 30: 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X‐Y, Zhou M, Yoo H, Pruneda‐Paz JL, Spivey NW, Kay SA, Dong X. 2015. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proceedings of the National Academy of Sciences, USA 112: 9166–9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY. 2016. TOC1‐PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis . Nature Communications 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Characterization of bZIP63 mutants.
Fig. S2 bzip63‐2 mutant has a reduced leaf area and growth rate.
Fig. S3 bZIP63 binds to energy stress‐responsive genes.
Fig. S4 bzip63‐2 starch degradation pattern under SD conditions.
Fig. S5 Circadian phase of KIN10‐induced energy‐stress responsive genes.
Fig. S6 Starch availability in SD vs 12 h : 12 h photoperiods.
Fig. S7 bZIP63 binds to cell wall modification gene.
Table S1 Up‐ and downregulated genes in bzip63‐2 plants compared with the WT.
Table S2 Genes related to circadian clock, starch degradation and energy stress deregulated in bZIP63 mutants.
Table S3 Relative metabolite content at EN in leaves of bzip63‐2 as compared to Ws.
Table S4 List of primers used for quantification of the mRNA levels and the immunoprecipitated sequences by qRT‐PCR.
Table S5 Circadian parameters calculated from two 24‐h cycles under LL conditions.
Table S6 Circadian oscillator binding in downregulated genes in bzip63‐2 that are induced by KIN10 overexpression.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


