Fig. 4.
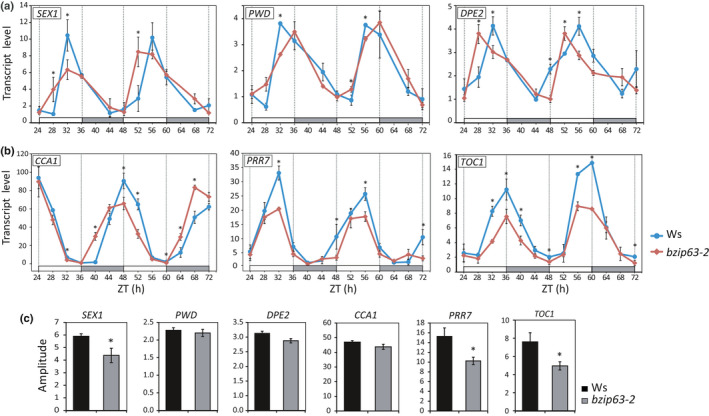
bZIP63 modulates the oscillatory pattern of circadian clock and starch degradation related genes. Transcript oscillatory pattern of starch degradation related genes (a), and circadian clock genes (b), are altered in bzip63‐2 as compared to Ws. 30‐d‐old plants were entrained under 12 h : 12 h, light : dark photoperiod and released into free‐running conditions (LL) for 3 d. The x‐axis represents the time elapsed after release of the plants into LL, where subjective day (ZT 24–36; 48–60) and subjective night (ZT 36–48; 60–72) are identified by white and gray bars on the x‐axis, respectively. Transcript oscillation amplitude of starch degradation‐related genes STARCH EXCESS1 (SEX1), PHOSPHOGLUCAN, WATER DIKINASE (PWD) and DISPROPORTIONATING ENZYME 2 (DPE2) and the circadian clock genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), PSEUDO‐RESPONSE REGULATOR 7 (PRR7) and TIMING OF CAB EXPRESSION 1 (TOC1) under LL (c). The data were obtained from two independent experiments, with three biological replicates in each time point, and each replicate consisted of the whole rosette collected from three plants (two‐tailed Student’s t‐test; n = 9; *, P < 0.05). Values are means, and error bars indicate standard deviation.
