Fig. 5.
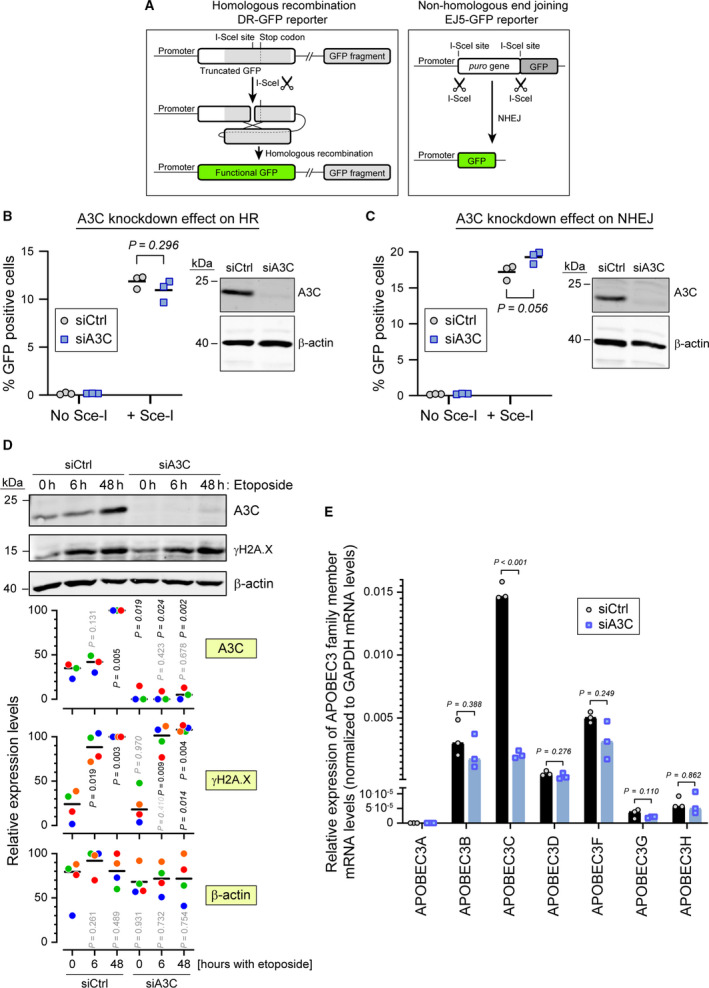
A3C is not required for efficient double‐strand DNA break repair. (A) Schematic representation (not to scale) of the DRGFP reporter (left) and EJ5GFP reporter (right). The DRGFP construct bears a cDNA encoding the full‐length GFP that contains an I‐SceI restriction site disrupting the reading frame and leading to a premature stop codon. This cDNA is followed by a 5′‐ and 3′‐truncated GFP fragment that does not contain the I‐SceI restriction site. When cells expressing this reporter are transduced with a lentivirus encoding the I‐SceI enzyme, the reporter is cleaved at the I‐SceI site. The ensuing DNA repair by HR removes the termination codon in the full‐length cDNA allowing for the production of a functional GFP (green). The EJ5GFP bears a puromycin resistance gene flanked by two I‐SceI restriction sites. This gene is followed by a GFP‐encoding cDNA. In cells expressing this reporter, removal of the puromycin resistance gene is achieved by transduction with an I‐SceI encoding lentivirus. The ensuing NHEJ‐mediated repair brings the promoter and the GFP cDNA close together, allowing for GFP production. (B, C) U2OS‐DRGFP (panel B) or U2OS‐EF5GFP (panel C) cells were transfected with either control siRNA or siRNA targeting A3C for 48 h and then transduced or not with a lentivirus carrying the ISce‐I endonuclease. Forty‐eight hours later the percentage of GFP positive cells was quantified by flow cytometry. Statistical analysis: unpaired Student’s t test. The right side of each panel shows the efficiency of A3C knockdown as assessed by western blotting. (D) Wild‐type U2OS cells were transfected with control siRNA (siCtrl) or siRNAs targeting A3C (siA3C) for 72 h, and then treated with 5 µm etoposide for the indicated time periods. Cell lysates were used for western blotting, with the indicated antibodies. The quantitation shown below the western blots are normalized to the ‘siCtrl, 48 h‘ values for A3C and γH2A.X and to the maximum values for β‐actin. Data points with the same color are derived from the same independent experiment. The black horizontal bars correspond to the median. The p‐values were obtained through paired t tests. For A3C and γH2A.X: nonitalicized and italicized values were those obtained by comparison with the corresponding ‘0 h‘ condition and between the ‘siCtrl‘ and ‘siA3C’ conditions at a given time point, respectively. For β‐actin, the p‐values were obtained by comparison with the ‘siCtrl, 0 h‘ condition. Shaded P values are those > 0.05. (E) A3C in U2OS cells was knocked down using a pool of 30 different siRNAs (siA3C). As a negative control, the cells were also treated with a nontargeting control siRNA pool (siCtrl). The specificity of the knock down was assessed by quantitating the mRNA levels of APOBEC3 family members.
