Abstract
Candida dubliniensis phenotypically mimics Candida albicans in its microbiological features; thus, its clinical characteristics have yet to be fully elucidated. Here we report the case of a 68-year-old Japanese man who developed C. dubliniensis fungemia during treatment for severe coronavirus disease 2019 (COVID-19). The patient was intubated and received a combination of immunosuppressants, including high-dose methylprednisolone and two doses of tocilizumab, as well as remdesivir, intravenous heparin, and ceftriaxone. A blood culture on admission day 11 revealed Candida species, which was confirmed as C. dubliniensis by mass spectrometry. An additional sequencing analysis of the 26S rDNA and ITS regions confirmed that the organism was 100% identical to the reference strain of C. dubliniensis (ATCC MYA-646). Considering the simultaneous isolation of C. dubliniensis from a sputum sample, the lower respiratory tract could be an entry point for candidemia. Although treatment with micafungin successfully eradicated the C. dubliniensis fungemia, the patient died of COVID-19 progression. In this case, aggressive immunosuppressive therapy could have caused the C. dubliniensis fungemia. Due to insufficient clinical reports on C. dubliniensis infection based on definitive diagnosis, the whole picture of the cryptic organism is still unknown. Further accumulation of clinical and microbiological data of the pathogen is needed to elucidate their clinical significance.
Keywords: Candida dubliniensis, Candidemia, COVID-19, Sequencing analysis
1. Introduction
Among the various known fungal infections, invasive candidiasis is the most frequent and serious, with candidemia as a representative disease [1]. With progress in medical intervention, Candida species have become common pathogens causing bloodstream infections and a high mortality rate among patients in intensive care units [2]. Clinically common Candida species include Candida albicans, followed by Candia glabrata, Candia tropicalis, Candia parapsilosis, Candia krusei, Candida guilliermondii, and Candida lusitaniae [3].
Candida dubliniensis was first documented in 1995 after its isolation from an oral sample of HIV-infected individuals in Dublin, Ireland [4]. Thereafter, this organism has been isolated from a variety of clinical samples from various countries worldwide. However, C. dubliniensis is clinically a less common species among approximately 150 Candida species [3], accounting for 0.1–1% of Candida species isolated from humans. It is a representative isolate of cryptic Candida species, formerly considered C. albicans because of its phenotypical similarity in microscopic morphology and production of pseudohyphae, true hyphae, and chlamydospores [5,6]. Previous studies demonstrated that nearly 2–3% of C. dubliniensis isolates may be misidentified as C. albicans [[7], [8], [9], [10]]. Although rare, C. dubliniensis potentially causes invasive infections [11,12]; thus, its clinical features should be documented in greater detail by the accumulation of clinical cases. Here we describe a case of C. dubliniensis fungemia in a fatal case of novel coronavirus disease 2019 (COVID-19).
2. Case report
A 68-year-old Japanese man was emergently transferred to our hospital with acute respiratory failure symptoms that had deteriorated over the previous 5 days. His medical history included myocardial infarction and a right lower lung lobe resection for lung cancer. On arrival, he was alert and his vital signs were as follows: body temperature, 37.0 °C; blood pressure, 138/107 mmHg; pulse, 85 beats/min; respiratory rate, 30/min; and oxygen saturation, 92% on 6 L/min of oxygen. In the physical examination, lung auscultation revealed bilateral course crackles, while laboratory testing showed elevated levels of C-reactive protein (7.03 mg/dL; normal, <0.14 mg/dL) and d-dimer (19.2 μg/mL; normal, <1.0 μg/mL). Lung computed tomography detected bilateral non-segmental ground-glass opacity. The patient was positive for PCR testing of severe acute respiratory syndrome coronavirus-2 and was hospitalized with a diagnosis of severe COVID-19.
He immediately received combination treatment consisting of methylprednisolone (250 mg/day for 3 consecutive days), remdesivir, tocilizumab (8 mg/kg), anticoagulant therapy with intravenous heparin, and ceftriaxone (2 g/day). However, his respiratory condition progressively deteriorated, and he was moved to an intensive care unit on admission day 2 and intubated the following day. On day 4, an additional dose of tocilizumab (8 mg/kg) was administered, and corticosteroid therapy was continued prednisolone (80 mg/day). On day 8, the antimicrobial therapy was empirically changed to meropenem and micafungin (150 mg/day).
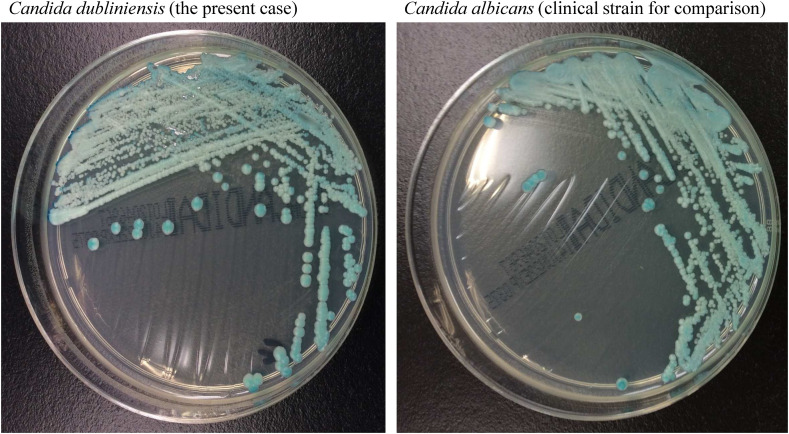
A blood culture on day 11 was positive for a yeast-like fungus. We sub-cultured the organism on BBL™ CHROMagar™ Candida II Medium (BBL, Beckton Dickinson GmbH, Heidelberg, Germany) and found that its morphological appearance was indistinguishable from that of C. albicans (Fig. 1 ). However, mass spectrometry (MALDI Biotyper; Bruker Daltonics Inc., Billerica, MA, USA) identified the organism as C. dubliniensis, with score values of 1.561 on incubation day 1, 1.740 on day 2, and 1.990 on day 3. C. dubliniensis was also isolated from a sputum sample on the same day. The central venous line was removed, which was negative for the pathogen, and the administration of micafungin was continued. Two weeks of antifungal therapy successfully eradicated the infection without persistent candidemia; however, his general condition progressively deteriorated, and he passed away after 5 weeks of hospitalization.
Fig. 1.
Morphological comparison of Candida dubliniensis and Candida albicans.
Candida species were cultured on BBL™ CHROMagar™ Candida II medium (BBL, Beckton Dickinson GmbH, Heidelberg, Germany) for 2 days at 35 °C.
2.1. Fungal identification
During the clinical course, we detected C. dubliniensis in blood and sputum samples. Minimum inhibitory concentrations of antifungal agents were as follows: amphotericin, 0.5 μg/mL; fluconazole, 0.06 μg/mL; itraconazole, 0.03 μg/mL; voriconazole, ≤0.015 μg/mL; micafungin, 0.03 μg/mL; and caspofungin, 0.25 μg/mL by the microdilution method using Dry Plate ‘Eiken’ (Eiken Chemical Co., Ltd, Tokyo, Japan). The blood-origin isolate was transferred for further in-depth analysis.
We performed direct polymerase chain reaction of the 26S rDNA and ITS regions. The primers for the targeted genes were as follows: NL1 (5′-GCATATCAATAAGCGGAGGAAAAG -3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) for 26S rDNA; and forward (5′-TCCGTAGGTGAACCTGCGG-3′) and reverse (5′-TCCTCCGCTTATTGATATGC-3′) for the ITS region [13]. The sequence data were analyzed using the Basic Local Alignment Search Tool (BLAST). The concordance rates for 26S ribosomal DNA (577 bp) and ITS (480 bp) were both 100% of the reference strains of C. dubliniensis (ATCC MYA-646). Therefore, the organism was identified as C. dubliniensis.
3. Discussion
Here we reported a clinical case of C. dubliniensis fungemia in a patient with severe COVID-19 who received two doses of tocilizumab in combination with high-dose corticosteroids. Although the infectious focus was unclear in the present case, considering the isolation of C. dubliniensis from the sputum, a lower respiratory tract infection was possible. Aggressive immunosuppressive treatment is recommended for patients with severe COVID-19 [14], and opportunistic infections, including candidemia, are increasingly reported in such cases [15,16].
C. dubliniensis is an infrequent and cryptic pathogen; thus, its clinical features remain to be elucidated. Most C. dubliniensis isolates have been isolated from the respiratory tract [10], especially oral samples [17,18]; however, recent reports have suggested that rare fungi can cause invasive infections as well [11,12]. Although rare, one case of C. dubliniensis fungemia during the treatment of COVID-19 has been described in the literature [19]. Similar to our case, that patient received immunosuppressive therapy consisting of dexamethasone and baricitinib, which was assumed to have contributed to the development of the C. dubliniensis fungemia. Unfortunately, the details of the fungal identification in that case were not fully discussed.
We identified the causative organism as C. dubliniensis based on a genetic investigation of the 26S rDNA and ITS regions. More specifically, the hyphal wall protein 1 gene polymorphism is currently available for species differentiation [20,21]. However, these methods are not accessible everywhere. A recent study corroborated that colony color on HiCrome candida differential agar after 72 h of incubation or growth on xylose-based agar medium after 48 h incubation is an easy, accurate, and cost-effective method of differentiating C. dubliniensis from C. albicans [22]. A mass spectrometry identification approach of C. dubliniensis appears reliable [23,24]; however, more database reviews are warranted [25,26]. Notably, nearly one-fifth of C. dubliniensis isolates showed mass spectrometry scores of <1.7 [10]. However, our data suggest that an inadequate incubation period may result in lower score values.
C. dubliniensis was previously considered an azole-resistant fungus. However, recent studies reported a good susceptibility of C. dubliniensis to azole-class antifungal drugs [21,27,28]. In fact, the COIVD-19–associated C. dubliniensis fungemia case was successfully treated with fluconazole [19]. However, an in vitro experiment suggested that fluconazole resistance may be induced in C. dubliniensis [29].
In conclusion, we herein presented a rare case of C. dubliniensis fungemia in a patient with severe COVID-19 in which the causative pathogen was genetically identified. C. dubliniensis is relatively new species and possibly overlooked in the laboratory because of its microbiological similarity to C. albicans. Thus, clinical features of C. dubliniensis infection do not appear to be fully elucidated. More clinical and microbiological data are required to increase our understanding of the cryptic pathogen.
Authorship statement
All authors met the ICMJE authorship criteria. AK and HH wrote the first draft of the manuscript. AK and IK analyzed the pathogens. YN and KH contributed to the clinical management of the patients. AH and FO supervised the study. All authors approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work in this paper.
Acknowledgments
We would like to thank Editage (www.editage.jp) for the English language editing.
References
- 1.Zaoutis T.E., Argon J., Chu J., Berlin J.A., Walsh T.J., Feudtner C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–1239. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 2.Eggimann P., Garbino J., Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/S1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 3.Johnson E.M. Rare and emerging Candida species. Curr Fungal Infect Rep. 2009;3:152–159. doi: 10.1007/s12281-009-0020-z. [DOI] [Google Scholar]
- 4.Coleman D.C., Sullivan D.J., Bennett D.E., Moran C.P., Barry H.J., Shanley D.B. Candidiasis: the emergence of a novel species. Candida dubliniensis. AIDS. 1997;11:557–567. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan D.J., Westerneng T.J., Haynes K.A., Bennett D.E.C.D. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiol. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 6.Jan A., Bashir G., Fomda B.A., Khangsar D.A.U., Manzoor M., Kohli A., et al. Hypertonic xylose agar medium: a novel medium for differentiation of Candida dubliniensis from Candida albicans. Indian J Med Microbiol. 2017;35:518–521. doi: 10.4103/ijmm.IJMM_17_216. [DOI] [PubMed] [Google Scholar]
- 7.Odds F.C., Van Nuffel L., Dams G. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J Clin Microbiol. 1998;35:2869–2873. doi: 10.1128/jcm.36.10.2869-2873.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly S.M., Sullivan D.J., Shanley D.B., Coleman D.C. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–1882. doi: 10.1099/13500872-145-8-1871. [DOI] [PubMed] [Google Scholar]
- 9.Tamura M., Watanabe K., Mikami Y., Yazawa K., Nishimura K. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: analysis reveals a new genotype of C. albicans with group I intron. J Clin Microbiol. 2001;39:4309–4315. doi: 10.1128/JCM.39.12.4309-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahelová M., Růžička F. Methods of Candida dubliniensis identification and its occurrence in human clinical material. Folia Microbiol (Praha) 2017;62:401–408. doi: 10.1007/s12223-017-0510-2. [DOI] [PubMed] [Google Scholar]
- 11.Lai C.C., Tsai H.Y., Chang T.C., Hsueh P.R. Catheter-related fungemia caused by Candida dubliniensis. J Microbiol Immunol Infect. 2013;46:306–308. doi: 10.1016/j.jmii.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Bosco-Borgeat M.E., Taverna C.G., Cordoba S., Isla M.G., Murisengo O.A., Szusz W., et al. Prevalence of Candida dubliniensis fungemia in Argentina: identification by a novel multiplex PCR and comparison of different phenotypic methods. Mycopathologia. 2011;172:407–414. doi: 10.1007/s11046-011-9450-6. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman C.P., Robnett C.J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5’ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartoletti M., Azap O., Barac A., Bussini L., Ergonul O., Krause R., et al. ESCMID COVID-19 living guidelines: drug treatment and clinical management. Clin Microbiol Infect. 2022;28:222–238. doi: 10.1016/j.cmi.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto K., Nakamura K., Hagiya H., Otsuka F. Candidemia in COVID-19 treated with corticosteroids and tocilizumab. Clin Case Rep. 2021 doi: 10.1002/ccr3.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arastehfar A., Ünal N., Hoşbul T., Alper Özarslan M., Sultan Karakoyun A., Polat F., et al. Candidemia among coronavirus disease 2019 patients in Turkey admitted to intensive care units: a retrospective multicenter study. Open Forum Infect Dis. 2022;9:ofac078. doi: 10.1093/ofid/ofac078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesee S., Samaranayake L., Roytrakul S., Paaopanchon C.P.P. Prevalence and susceptibility profiles of oral yeast species isolated from a healthy adult Thai cohort. Arch Oral Biol. 2022;138 doi: 10.1016/j.archoralbio.2022.105415. [DOI] [PubMed] [Google Scholar]
- 18.Jabri B., Iken M., Ait-Ou-Amar S., Rida S., Bouziane A.E.O. Candida albicans and Candida dubliniensis in periodontitis in adolescents and young adults. Internet J Microbiol. 2022;2022 doi: 10.1155/2022/4625368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alataby H., Atemnkeng F., Bains S.S., Kenne F.M., Diaz K., Nfonoyim J. A COVID-19 case complicated by Candida dubliniensis and Klebsiella pneumoniae -Carbapenem-Resistant enterobacteriaceae. J Med Cases. 2020;11:403–406. doi: 10.14740/jmc3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salehipour K., Aboutalebian S., Charsizadeh A., Ahmadi B.M.H. Differentiation of Candida albicans complex species isolated from invasive and non-invasive infections using HWP1 gene size polymorphism. Curr Med Mycol. 2021;7:34–38. doi: 10.18502/cmm.7.2.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shokoohi G., Javidnia J., Mirhendi H., Rasekh-Jahromi A., Rezaei-Matehkolaei A., Ansari S., et al. Molecular identification and antifungal susceptibility profiles of Candida dubliniensis and Candida africana isolated from vulvovaginal candidiasis: a single-centre experience in Iran. Mycoses. 2021;64:771–779. doi: 10.1111/myc.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan A., Bashir G., Altaf I., Fomda B.A., Hamid S.J.K. Evaluation of various phenotypic methods for differentiation of Candida dubliniensis from Candida albicans. J Med Microbiol. 2021;193 doi: 10.1016/j.mimet.2021.106400. [DOI] [PubMed] [Google Scholar]
- 23.Lee H.S., Shin J.H., Choi M.J., Won E.J., Kee S.J., Kim S.H., et al. Comparison of the bruker biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann Lab Med. 2017;37:223–230. doi: 10.3343/alm.2017.37.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancini N., De Carolis E., Infurnari L., Vella A., Clementi N., Vaccaro L., et al. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass Spectrometry systems for identification of yeasts of medical importance. J Clin Microbiol. 2013;51:2453–2457. doi: 10.1128/JCM.00841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teke L., Barış A., Bayraktar B. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for non-albicans Candida and uncommon yeast isolates. J Microbiol Methods. 2021;185 doi: 10.1016/j.mimet.2021.106232. [DOI] [PubMed] [Google Scholar]
- 26.Korem M., Cohen M.J., Michael-Gayego A., Castiel D., Assous M.V., Amit S. Misidentification of Candida dubliniensis isolates with the VITEK MS. J Med Mycol. 2021;31 doi: 10.1016/j.mycmed.2020.101107. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan D.J., Moran G.P., Coleman D.C. Candida dubliniensis: ten years on. FEMS Microbiol Lett. 2005;253:9–17. doi: 10.1016/j.femsle.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller M.A., Diekema D.J. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J Clin Microbiol. 2004;44:4419–4431. doi: 10.1128/JCM.42.10.4419-4431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borg-von Zepelin M., Niederhaus T., Gross U., Seibold M., Monod M., Tintelnot K. Adherence of different Candida dubliniensis isolates in the presence of fluconazole. AIDS. 2002;16:1237–1244. doi: 10.1097/00002030-200206140-00005. [DOI] [PubMed] [Google Scholar]