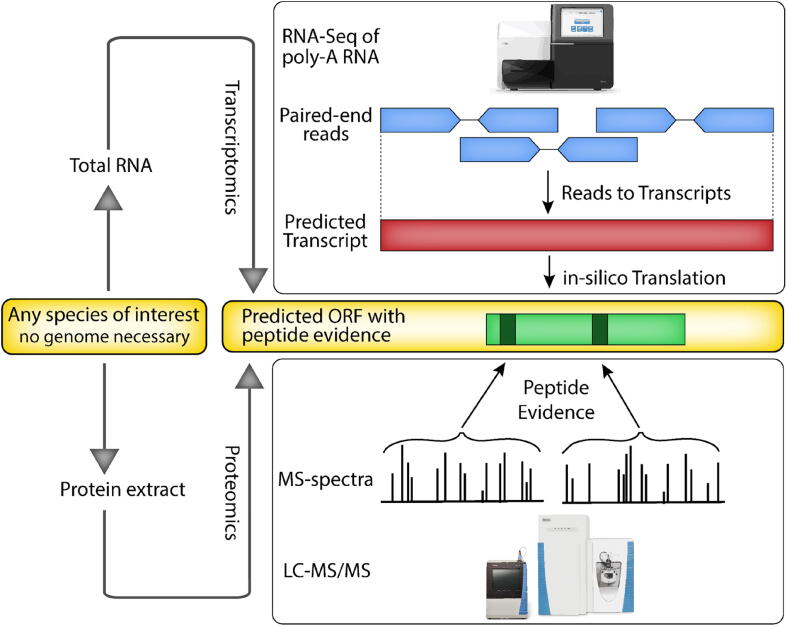
Fig. 3.
General outline of the PTA (Proteo-Transcriptomics Assembly) approach. RNA-sequencing data of all poly-adenylated RNA molecules of any species of any cell origin is used for transcript assembly in which individual reads are concatenated into potential full-length transcript contigs. The predicted transcript contigs are then in-silico translated into predicted protein sequences in all possible frames. These predictions are used to find potential open reading frames taking important features of common protein coding transcripts (such as a Methionine start and an in-frame stop codon) into consideration. In parallel the proteome of the same sample used for RNA-sequencing is measured with a high-resolution mass spectrometer. The mass spectrometer first records the mass/charge (m/z) of each peptide ion and then selects the peptide ions individually to obtain sequence information via MS/MS. Peptide fragmentation spectra are matched to in silico generated peptide fragmentation patterns. The ultimate result of the process are transcript contigs that were validated by the presence of peptides and hence represent a set of high confidence protein coding transcripts.