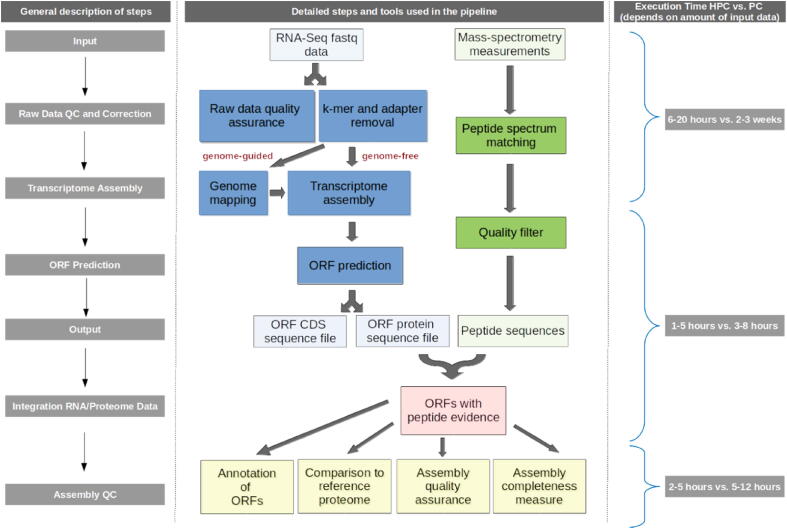
Fig. 4.
General outline of the PTA workflow. In blue: RNA-Seq data preparation steps include 1. the validation of sufficient quality of the sequencing data (FastQC[52], fastqp [53], fastq-stats [54]); 2. raw RNA-Seq reads correction and adapter removal (Rcorrector[55], QuorUM [56], specialized scripts from TranscriptomeAssemblyTools (FilterUncorrectablePEfastq.py); TrimGalore (a wrapper around Cutadapt [57] and FastQC [52]); 3. Mapping of reads to a reference genome for the genome-guided mode (STAR[58], Bowtie2 [59], BWA [60], Hisat2 [61], TopHat2 [62]); 4. Transcriptome assembly (Trinity[20], [21], Oases [22], Trans-ABySS [19], SOAPdenovo-Trans [24], IDBA-Tran [23], Bridger [26], BinPacker [27], Shannon [25], SPAdes-sc [28], SPAdes-rna [28]); 5. Identification of candidate coding regions within reconstructed transcript sequences from the previous step (TransDecoder[21], FrameD [63], GeneMarkS [64]). In green: mass spectrometry spectra processing and filtering (MaxQuant[65], ProteomeDiscoverer (Thermo Scientific), FragPipe [66], MS-GF+ [67]). In red: The predicted ORF protein sequences will be used as search space for the identified peptides extracted from MS/MS spectra. In yellow: ORFs with peptide evidence can be functionally annotated (Trinotate[68], blast2GO [69], annot8r [70], Annoscript2 [71]). Newly established annotations can be compared with current annotations e.g., from UniProt and Ensembl (blastp[72], DIAMOND [73]), checked for assembly quality standards (TransRate[40], rnaQUAST [39], Detonate [41]) and examined for proteome completeness (BUSCO[42]). Programs that can be used for the individual steps are listed, while the ones that were tested to work well and deliver satisfactory results in our hands are bolded. The list, though being comprehensive, is not intended to be complete. Beyond the tools listed, alternative tools that may work equally well may exist or being developed. The right panel depicts the computation times of the different steps compared between High-Performance-Computing machines and strong tabletop PCs. The times are only representative, based on the tools marked bold, and depend on the amount of raw data processed and the underlying computing architecture. Execution time may vary for alternative tools used for the individual steps.