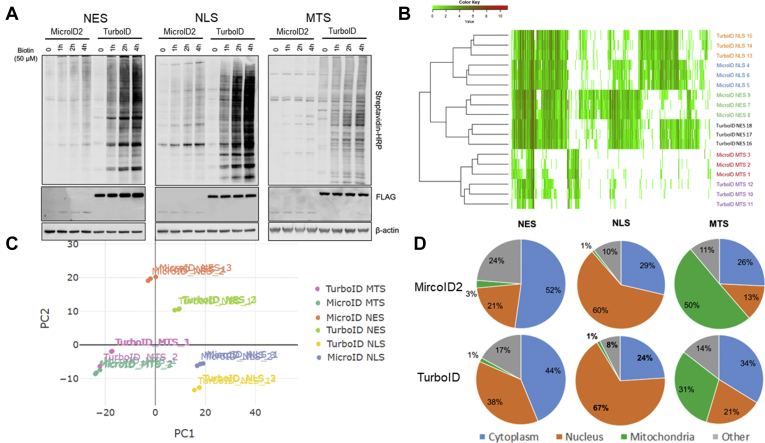
Fig. 8.
Proteomics of subcellular organelles with MicroID2.A, biotinylation activity of stably integrated MicroID2 and TurboID constructs. Following stable integration of the biotin ligase constructs using the sleeping beauty transposon system, we treated cells with biotin for the indicated times. After cell lysis, we performed Western blotting to examine MicroID/TurboID construct expression (FLAG) and biotinylation (streptavidin-488). Representative of two independent experiments. B, heat map of all proteins identified using the Prostar proteomics GUI (26). C, MDS plot was generated from the top 500 proteins identified using the TCC GUI package (27). D, stable MicroID2 and TurboID BEAS-2B cell lines were treated with exogenous biotin for 3 h (MicroID2) or 1 h (TurboID). After harvesting the cells, we pulled down biotinylated proteins with streptavidin–HRP and submitted for mass spectrometry. Un-normalized spectral counts present in at least two samples and averaging to three peptides per group were included in the analysis. Unique proteins identified and mapped to subcellular organelles: MicroID-NES = 1639; MicroID-NLS = 1686; MicroID-MTS = 334; TurboID-NES = 1700; TurboID-NLS = 1635; and TurboID-MTS = 379. n = 3 samples per group. UniProt IDs were utilized to assign subcellular location using the “Retrieve/ID Mapping” tool from the UniProt database. GUI, graphical user interface; HRP, horseradish peoxidase.