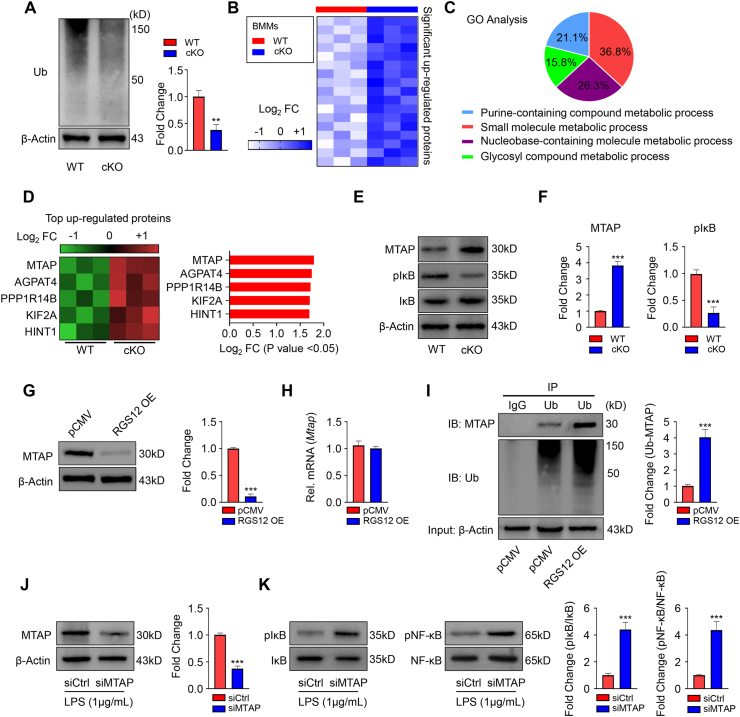
Figure 4.
The loss of RGS12 inhibits ubiquitination and promotes molecule metabolic process. (A) Immunoblot showed the Ub protein in WT and RGS12 cKO BMMs. Quantitative data showed the relative levels of Ub. A t test showed significant differences between the two groups, ∗∗P < 0.01 versus WT. Data are representative of three separate experiments. (B) Heatmap depicting the proteins which were up-regulated in RGS12 KO BMMs as compared to control cells. Optimized cutoff thresholds for significantly altered proteins was set at 1.3 log2-transformed ratios and P < 0.05, (n = 3). (C) Gene ontology (GO) enrichment analysis to identify biological processes corresponding to the significantly altered proteins. The percentage of significant biological processes was shown in the pie chart with various colors. (D) Heatmap and bar graph depicting the top 5 significant up-regulated proteins as depicted in (B). (E) Immunoblot showed the MTAP, pIκB, IκB, and β-Actin protein levels in WT and RGS12 cKO BMMs. (F) Quantitative data showed the MTAP and pIκB expression as depicted in (E). A t test showed significant differences between the two groups, ∗∗P < 0.001 versus WT. Data are representative of three separate experiments. (G) RAW264.7 cells were transfected with pCMV control and pCMV-RGS12 plasmids (RGS12 OE) for 48 h. The MTAP protein levels were detected by Western blot. Note that the overexpression of RGS12 promotes the degradation of MTAP in macrophages. ∗∗∗P < 0.001 versus pCMV group, (n = 3). (H) Gene expression levels of MTAP in the macrophages as described in (G). Data are reported as means ± SEM (n = 5). (I) RAW264.7 cells were transfected with pCMV or pCMV-RGS12 plasmids for 48 h with Lipofectamine 3000 (ThermoFisher, US). The cell lysates of those cells were incubated with anti-Ub or control immunoglobulin G (IgG) antibody, and bound protein was examined by Western blotting with the corresponding antibodies. The level of Ub associated MTAP (Ub-MTAP) was analyzed. Note that the overexpression of RGS12 enhanced the combination of Ub and MTAP. ∗∗∗P < 0.001 versus pCMV group, (n = 3). (J) The BMMs from WT mice were transfected with siCtrl and siMTAP for 24 h and treated with LPS (1 μg/mL) for 24 h. The MTAP protein level was determined by Western blot. Quantitative analysis of the MTAP level was shown on the right panel. The data are represented as means ± SEM. ∗∗∗P < 0.001, n = 3. (K) The levels of pIκB, IκB, pNF-κB, and NF-κB in BMM lysis were detected by immunoblotting as described in (J). Quantitative analyses were shown on the right panel. Note that reduced MTAP enhanced the expression of pNF-κB and pIκB. ∗∗∗P < 0.001, n = 3.