Abstract
Macrophages are dominant innate immune cells. They demonstrate remarkable heterogeneity and plasticity that are essential for homeostasis and host defense. The heterogeneity of tissue macrophages is shaped by the ontogeny, tissue factors, and environmental signals, a pattern in a tissue-associated latitudinal manner. At the same time, macrophages have long been considered as mainly plastic cells. These cells respond to stimulation quickly and in a stimulus-specific way by utilizing a longitudinal cascaded activation, including coordination of signal transducer, epigenetic elements, and transcription factors, conclusively determine the macrophage phenotypes and functions. With the development of cutting-edge technologies, such as fate-mapping, single-cell transcriptomics, ipsc platform, nanotherapeutic materials, etc., our understanding of macrophage biology and the roles in the pathogenesis of diseases is much advanced. This review summarizes recent progress on the latitudinal and longitudinal regulation of tissue macrophages in inflammatory diseases. The latitudinal regulation covers the tissue macrophage origins, tissue factors, and environmental signals, reflecting the macrophage heterogeneity. The longitudinal regulation focuses on how multiple factors shape the phenotypes and functions of macrophage subsets to gain plasticity in inflammatory diseases (i.e., inflammatory bowel disease). In addition, how to target macrophages as a potential therapeutic approach and cutting edge-technologies for tissue macrophage study are also discussed in this review.
Keywords: Epigenetic regulation, Inflammatory disease, microRNA, Macrophage origin, Macrophage polarization, Single-cell RNA-sequencing, Tissue macrophage
Introduction
Macrophages are developed in different tissues, where they undergo phagocytosis, process foreign materials, clear debris, and secrete cytokines in response to activation signals.1 These functions are essential in the development, tissue homeostasis, and inflammation resolution.2 Among the brain, heart, liver, lung, intestine, spleen, kidney, etc., macrophages represent the most heterogeneous and plastic immune cell population.3 Therefore, clarifying the mechanisms behind the scenes is vital for understanding the association of tissue-specific macrophage and disease pathogenesis (Table 1). Latitudinal and longitudinal regulations are two-dimensional approaches. The latitudinal regulation covers the tissue macrophage origins, tissue factors, and environmental signals, reflecting the macrophage heterogeneity. The longitudinal regulation focuses on how multiple factors trigger signal cascade and shape the phenotypes and functions of macrophage subsets to gain plasticity in inflammatory diseases.4 With the cutting-edge technologies’ development, our understanding of macrophage biology and its roles in disease pathogenesis is advanced.5 This review provides informed knowledge of tissue-specific macrophages associated with disease pathophysiology, especially inflammation; it offers a perspective on therapeutic targets exploration (Table 1).
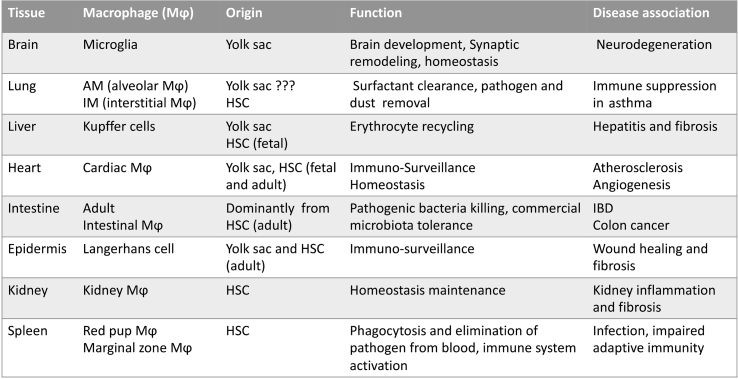
Table 1.
Basic information of various tissue macrophages.
HSC: hematopoietic stem cell.
The origins and characters of tissue macrophages
General information of tissue macrophage origins
Macrophages arise from the yolk sac and fetal liver progenitors during embryonic development; they continue to differentiate in different organs as heterogeneous, self-maintaining tissue-resident macrophages.3,6, 7, 8 After birth, blood monocytes from bone marrow are recruited to replenish tissue-resident populations and adapt to specific environmental cues like infection, inflammation, and metabolic perturbations.6,9, 10, 11 The mixed origins of tissue macrophages are varied in different tissues. Evidence showed that most murine tissue macrophages in the brain, epidermis, alveoli of the lung, and liver develop from the embryonic origin, which gives rise to fetal macrophages early during embryonic development.8 Differently, macrophages in the dermis, intestine, and peritoneum mainly rely on the monocyte-origin to fill the macrophage repertoire.12 In most tissues, the dual origins of macrophage co-exist, but the ratio varies. Tissue-resident macrophages differ in replication and turnover due to the difference of origins and locations, but they occupy the general macrophage functions such as phagocytosis ability, cytokines production, and host interaction, which are in an organ-specific manner.4 We highlight some tissues to describe the origin, phenotype, function, and disease association of these tissue macrophages (Fig. 1).
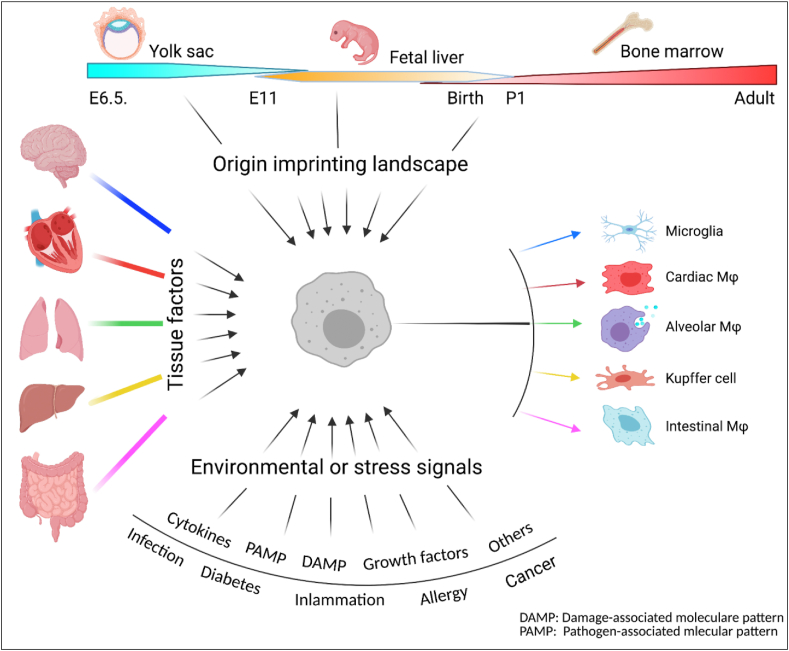
Figure 1.
Latitudinal heterogeneity of tissue macrophages. Multiple dimensional signals determine tissue macrophage heterogeneity in a latitudinal manner. Firstly, the origins imprint the macrophages with the ontogeny landscape; tissue signals then provide unique information associated with functions; macrophages in the niche finally sense the environment and stress signal to differentiate into a specific macrophage subset in homeostasis or disease condition.
Brain—microglia cell
Microglia location and functions
Microglia are tissue-resident macrophage-like innate immune cells in the central nerve system (CNS), they have memory-like functions to allow context-dependent responses.13 Though the microglial heterogeneity can be observed according to their morphology and density in specific brain compartments, the expression of cellular markers is still essential to define the microglia subpopulation and their associated functions. However, choosing the appropriate makers is very challenging because of the complexity of microglia markers. Usually, researchers use some general markers to combine with the resting or active markers to define the function-associated populations in the CNS. Moreover, resident CNS microglia share some majority of markers such as CD11b, F4/80, CX3CR1, CD45, and IBA-1 with monocyte-derived macrophages, which make it more difficult to distinguish the origins.
Microglia and neurodegeneration
As multifunctional cells, microglia interact with numerous other cells in the CNS, including neurons, astrocytes, and oligodendrocytes. In fact, genome-wide association studies (GWAS) have identified many risk genes of CNS disorders are expressed by microglia, including autism, Alzheimer's disease (AD), Parkinson's disease (PD), schizophrenia, and multiple sclerosis (MS). With the development of sequencing technologies, people uncovered those microglia are not just inactive bystanders of CNS pathologies but, rather, determinants of diseases. Besides, microglia primary defects are found to be critical in several monogenetically inherited microgliopathy that are associated with severe CNS pathologies.14 Therefore, microglia have emerged as a novel promising treatment target for numerous neuropsychiatric diseases.
Lung—AM and IM
Lung macrophage locations and functions
According to macrophage sites located, lung macrophages are defined as alveolar Mφs (AMs) and interstitial Mφs (IMs), two subsets.15 These subsets are shaped by their origins, microenvironment, and other tissue factors, which grant macrophages diverse characters and plasticity.16,17 Both AMs and IMs in mice show unique surface markers expression, which links subset phenotypes to functional complexity.
Lung AM and IM subsets
Dr. Misharin (2013) established a sequential gating methodology in flow cytometry to describe AM and IM.18 In a CD45+ population, AM cells express unique SiglecF+CD11b−CD11c+CD64+, whereas IM subset is CD11b+MHCII+CD11C+CD64+CD24- cells. Sorting these subsets can assist in further functional analysis.18, 19, 20 Recently, IMs are further clustered into several subsets depending on single-cell RNA data.21 Besides surface markers, AMs and IMs, can also be outlined by key transcription factors, such as PPAR-γ, PU.1, and C/EBPβ, associated with their differentiation from monocytic ancestors.22 Other transcriptional factors, such as BACH2, have also been important for developing and maintaining alveolar Mφs.23
Macrophages and lung inflammation
GM-CSF signaling is critical for alveolar Mφs development; GM-CSF−/− results in mature AMs loss, surfactant accumulation, and alveolar proteinosis.24 GM-CSF production needs TGF-β secreted by AMs. Loss of TGF-β leads to failure of development and maintenance of AMs throughout life. Unsurprisingly, Mφs have a range variety of functions in the lungs. To keep the lung homeostasis and maintain immune surveillance, they play essential roles in removing cellular debris, clearance of microbes, and inflammation resolution.16,25 The study recently found that impairing AM chemotaxis toward bacteria can induce redundant neutrophil recruitment, leading to inappropriate inflammation and injury. In a disease milieu, influenza A virus infection impaired AM crawling and significantly increased secondary bacterial co-infection.26 IMs expressing CD206+ (MHCIIlo CX3CR1lo), are peribronchial, involved in immunoregulation, wound healing, and repair, and are self-sustaining over an extended period.27
AMs and IMs have different capacities to self-renew over the organism's lifetime and recovery from inflammation and injury. Together, Mφs are the primary innate immune cells in the lung at homeostasis, with important host defense and immune modulation roles.
Liver—Kupffer cell and MoMφs
Liver macrophage origins and functions
Liver macrophages mainly contain Kupffer cells (KC), originating from yolk sac-derived precursors during embryogenesis, and monocyte-derived macrophages (MoMφs). Fundamentally, Kupffer cells have self-renewing, locally proliferating, and tolerogenic features; they receive signals from the local microenvironment that prompt their functional differentiation. MoMφs differentiated from bone marrow-derived circulating monocytes. During homeostasis, liver monocytes' contribution to hepatic macrophages is low, but monocytes are recruited mainly to inflammation areas after hepatic injury. MoMφs can gain Kupffer cells self-renew ability and certain characters if Kupffer cells are fully depleted; they are believed to be the significant contributors to the macrophage pool's replenishment.28
Liver Kupffer cell and MoMφ subsets
The expression of surface markers is a principal way to distinguish KCs and MoMφs cells. In mice, MoMφs cells are CD11b+, F4/80intermediate (int), Ly6C+, and CSF1R+ unique population; they are differentiated from circulating monocytes. In mouse models of liver diseases, hepatic MoMφs have Ly6Chigh and Ly6Clow, two main subpopulations. In mice, KCs are defined as CD11blow, F4/80high, and Clec4F+ population.29, 30, 31, 32 Besides the surface markers, Kupffer cells and MoMφs are also diverted on-location sites and functions. Kupffer cells are seeded along with sinusoidal endothelial cells; they continually clear gut-derived pathogens; they maintain iron and cholesterol homeostasis; notably, they promote immunological tolerance.33,34 Therefore, KCs replenish is an essential housekeeper in the steady-state, which is independent of BM-derived progenitors.35
Macrophages and liver injury
The phenotypes of liver macrophages are dynamically changing; Kupffer cells and MoMφs rapidly adapt their phenotypes in response to local signals, determining their ability to aggravate or cease liver injury. Interestingly, MoMφs can differentiate into fully functional KCs and restore the population of hepatic macrophages if Clec4F+ KCs population depleted, suggesting liver macrophages' plasticity.30,36 Furthermore, aside from the proliferation of KCs and the recruitment and differentiation of MoMφs, other cellular sources for hepatic macrophage replenishment such as infiltrating peritoneal GATA6+ macrophages might contribute to the pool of hepatic macrophages during liver injury. So far, the role of liver macrophages in the progression of viral hepatitis is still debated. Activated KCs, described by the upregulation of CD33 and CD163, gather in the portal tract during chronic HBV/HCV infection, emphasizing the importance of these cells in fighting viral hepatitis.37 Single-cell RNA sequencing technology has provided evidence that more specific macrophage subsets will be therapeutic targets for non-alcoholic fatty liver disease (NAFLD).38
Taken together, the comprehensive analysis on the origins, the phenotypes, and the functions of liver macrophages have suggested their heterogeneity and plasticity.39 The signals in the micro-environment include ROS, pattern recognized receptor (PAMP), damage-associated molecular patterns (DAMP), lipopolysaccharide (LPS), LTA, and β-glucan, microRNA and mitochondria RNA, etc. can target liver macrophages, which express a high density of PRRs, TLR, and NLRs. These diverse signal pathways are essential in liver disease pathogenesis, and liver macrophages promote the restoration of tissue integrity following liver injury or infection. However, they can also contribute to liver disease progression, including hepatitis, fibrosis, and cancer. Evidence from animal models and early clinical trials in humans indicate that targeting pathogenic liver macrophages might be a promising therapeutic approach in acute and chronic.
Intestinal macrophages—homeostasis and inflammation
Intestinal macrophage locations and functions
Principally, macrophages are present in all intestine layers, including laminar propria, submucosa, and the muscularis externa.40,41 The locations of intestine macrophages are associated with their functions.42,43 (1) In the muscularis externa, intestinal tissue-macrophages interact with neuron cells. As a piece of evidence, depletion of these embryonic-derived macrophages leads to increased neuronal apoptosis and altered morphology and abundance of blood vessels. These findings suggest that embryonic-derived macrophages influence enteric neurons and regulate essential intestinal functions, including secretion and motility. (2) Within the lamina propria, macrophages also play a vital role in maintaining the intestinal stem cells' function; they provide the WNT signal to the stem cell for the stemness. Meanwhile, macrophage ablation following CSF1R blockade affects Paneth cell differentiation and reduces Lgr5+ intestinal stem cells.44 (3) Laminar propria macrophages have important influences on T cell function. Though intestinal macrophages' dual origins are well accepted, the bone marrow-derived monocytes are still a major part of the lamina propria. In particular, the activation status of intestinal macrophages contributed to the activity and function of Tregs (regulatory T cells, via the production of IL-10)45 and Th17 cells (as a source of IL-1β).46 Therefore, macrophages maintain homeostasis in the gut through a combination of phagocytic and antibacterial functions and immune regulation.
Microbiota and Tim4+ intestinal macrophage subsets
As a unique organ that harbors microbiota, intestine macrophage phenotypes and functions are determined mainly by the microbiota and metabolites.12,47 A study found that butyrate, a bacterial fermentation product, induces differentiation of macrophages with microbicidal function, enhanced antimicrobial function and antimicrobial peptides, and inhibits HDAC3 to drive metabolic changes and antimicrobial function.48 Moreover, commensal microbiota was found to drive the functional diversification of colon macrophages CD11c+CD206intCD121b+ and CD11c−CD206hiCD121b− colon macrophage populations.49 This study highlights the importance of microbiota on the unique developmental and anatomical, and functional fates of colon macrophages. Recently, a group identified three transcriptionally distinct mouse gut macrophage subsets that segregate based on the expression of Tim4 and CD4. With challenging the current concept, Tim4+CD4+ gut macrophages self-maintained locally, while Tim4–CD4+ macrophages had a slow turnover from blood monocytes; indeed, Tim4–CD4– macrophages were the only subset with the high monocyte-replenishment rate currently attributed to gut macrophages.43 Remarkably, live microbiota is the major factor to sustain all these three macrophage subpopulations numbers; it suggested that: (1) Locally maintained subsets coexist with rapidly replenished monocyte-derived populations. This updated the previous prevailing paradigm that all macrophages in the adult mouse gut rapidly turn over from monocytes. (2) The significant intestine feature makes it different from other tissues, while locally maintained macrophage and monocyte-derived populations have collaborated in a live microbiome-dependent manner.
Macrophages in the pathogenesis and therapy of IBD
Inflammatory bowel disease (IBD) is a chronic disease developed from an acute onset of inflammation; it sometimes leads to life-threatening complications. Besides the Treg cells, macrophages play an essential role in colitis by secreting many cytokines and controlling tissue repair. During the pathogenesis of IBD, the macrophage sub-population component varied in the acute phase and chronic phase. Therefore, macrophage differentiation and polarization are essential events in IBD; identifying such a mechanism can develop a therapeutic target. A recent study found that extracellular matrix protein-1 (ECM1) was highly expressed in macrophages, particularly tissue-infiltrated macrophages, under inflammatory conditions, and ECM1 expression was significantly induced during IBD progression. They showed ECM1 deficiency in macrophages reduced sensitivity to dextran sodium sulfate-induced colitis with increased expression of arginase 1 and impaired M1 polarization by activating the STAT5 pathway. This finding showed that ECM1 has a vital function in endorsing M1 macrophage polarization, which is important for controlling inflammation and tissue repair in the intestine.50 This is strong evidence for targeting macrophage polarization as a potential strategy for IBD therapy. Recently, Brink's group found that anti-TNF therapy polarizes human and mouse macrophages towards CD206+ macrophages in an IL-10-dependent manner in vitro and in patients with Crohn's disease in vivo; these results illustrated the critical role of IL-10-induced repolarization of macrophages to a regulatory phenotype in the therapy of patients with IBD.51
Regulation mechanisms of macrophage differentiation and polarization
Macrophage activation and polarization are structured in longitudinal regulation, initiating from the origins, the tissue factors, the environmental and stress signals.52 These signals trigger a multiple-level cascade, including microRNAs and epigenetic markers modification, critical transcriptome shape, and phenotypes determination, and finally, define a specific proinflammatory or anti-inflammatory function macrophages in tissue (Fig. 2).53, 54, 55, 56
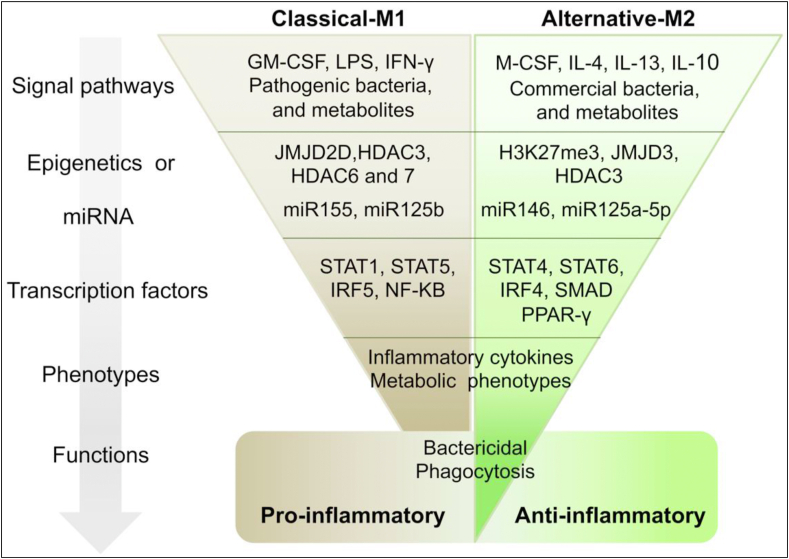
Figure 2.
Longitudinal regulation of macrophage polarization. Macrophage polarization is a multiple-level regulated event. In response to the environmental cues, microRNA and epigenetic modifier will work together to shape transcriptome then finally decide the phenotype and functions of polarized macrophages.
Macrophage differentiation signals and polarization spectrum
Macrophages ubiquitously exist in most tissues, but the phenotypes and functions are varied in different tissues. The tissue factor is vital for macrophage specificity. Intriguingly, a study found that surfactant protein A (SP-A) enhanced IL-4-dependent macrophage proliferation and activation in the lung. Also, in the peritoneal cavity and liver, C1q was essential for type 2 macrophage activation and liver repair following bacterial infection. This evidence suggested that SP-A and C1q, and the expression of their receptor, myosin 18A, are critical for tissue macrophage activation; they uncover the presence within different tissues of an amplification system needed for local macrophage responses and activation.57
Furthermore, intrinsic signal and environment signal are all controllers of macrophage phenotypes and functions. Recently, a study defines the role of the autophagy proteins Beclin 1 and FIP200 in facilitating quiescence of tissue-resident macrophages by limiting the effects of systemic interferon-γ. This evidence suggests that the autophagy signal pathway has a crucial function in maintaining macrophages' homeostasis status, dysfunction of autophagy resulting in bacterial infection's genetic susceptibility.58 Factors such as cytokines, growth factors, and microorganism-associated molecular patterns are thought to drive a transcriptional response that decides the phenotype and function of the macrophages based on the homeostasis or pathological context.
Two extremely polarized macrophages are classic activation macrophage (M1) and alternative activation macrophage (M2); they undergo pro- or anti-inflammation functions in infection, obesity, neurodegeneration, and cancer. Classifying two very extreme and opposite macrophage polarization statuses aims to match T cell responses in inflammation. However, it is widely recognized that the M1/M2 terminology has the limitation of describing the in vivo condition.59 The real world of macrophage polarization is in a spectrum pattern, reflects the complexity of macrophage differentiation and activation. However, M1 and M2 are still useful for in vitro work for pro-/anti-inflammatory function and inflammation study.60,61
MicroRNA and epigenetic regulation in macrophage polarization
microRNAs
Let-7c is the first identified miRNA determining macrophage polarization. Later, miR-125a-5p was found to have comparable characteristics to let-7c.62 This evidence offers a new understanding of the macrophage polarization mechanism. As two classical representatives, miR-155 and MiR-146a were well studied in driving M1 and M2 polarization, separately.63,64 Up to date data suggested that M1 polarization is also supported by other microRNAs, such as miR-181, miR-451, miR-720, miR-127, and miR-21. In particular, miR-127 and miR-125b, which have been revealed to target Bcl6 and IRF4, respectively, directly drive the pro-inflammatory cytokines' expression. Also, overexpression of miR-720 diminished the expression levels of GATA3, a transcription factor important in M2 macrophage polarization, therefore causing the inhibition of M2 polarization. Interestingly, some miRNA clusters miR-23a/27a/24-2 cluster are associated with M2 polarization. Therefore, miRNAs are part of the regulation cascade, miRNA-associated targets, and pathways that are promising to investigate macrophage biology and disease pathogenesis.
Histone modifications
Overall, the epigenetic landscape is the sum of three elements: (1) chromatin modifications; (2) pattern of DNA methylation; and (3) promoters and enhancers region pre-bound proteins. During the cell differentiation, origin and environment signal have first shaped the landscape.65,66 The subsequent stimulation and acute signal will further determine accessibility for binding signaling transcription factors such as nuclear factor (NF)-κB, SMAD, STATs, and PPARr, etc., during macrophage polarization.67, 68, 69, 70
The epigenetic study of macrophage polarization to date has mainly focused on histone modifications, which can be broadly divided into positive and negative marks that promote or suppress transcription, respectively. Histone tails modification decides the chromatin accessibility, which dynamically determines the gene expression profile. The lysine residues of histones H3 and H4 can be chemically modified by adding methyl or acetyl groups. Almost all the acetylation sites induced active transcription; as histone tails acetylation leads to a more open chromatin structure and is feasible for transcription factor binding to the regulatory regions; it increases the gene transcription (Fig. 3). Bromodomain-containing protein 4 (BRD4) is a typical ‘reader’ protein for recruiting additional transcriptional complexes.71 However, methylation's active or repression effect depends on lysine and arginine sites formats, including mono-methylation, di-methylation, or tri-methylation (Fig. 3). For example, H3K9me2 and H3K9me3 are associated with closed, compacted chromatin and suppress gene transcription by recruiting proteins such as heterochromatin protein 1 (HP1).
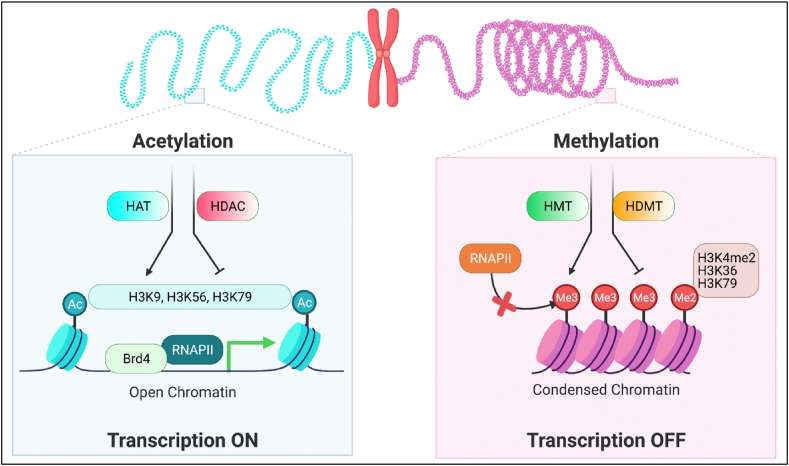
Figure 3.
Histone modification on the transcription of macrophages. Histone modification decides the status of chromatin. H3 tail acetylation at the promoter and enhancer regions will open the chromatin and increase TFs accessibility to promote gene transcription. On the opposite, H3 methylation will form condensed chromatin, which represses the gene transcription. HAT: Histone acetylation; HDAC: Histone deacetylation; HMT: Histone methylation transfer enzyme; HDMT: Histone demethylation transfer enzyme.
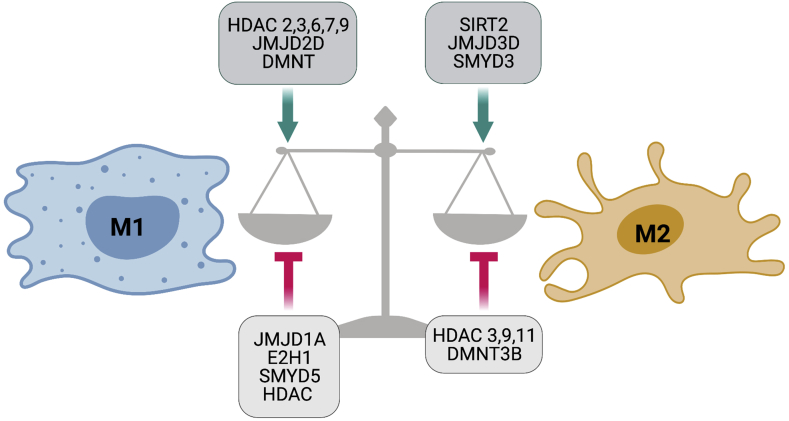
Histone modification enzymes decide the epigenetic regulation status during macrophage polarization to drive the polarized gene profile (Fig. 4). In particular, some HDAC, JMJD2D, and DMNT were induced in M1 macrophages in response to TLR stimulation, indicating a role of chromatin remodeling in inflammatory gene expression. Moreover, transcriptional activation induced during M2 polarization is commonly associated with SIRT2, JMJD3D, and SMYD3, etc. In detail, the JMJD3-IRF4 axis is necessary for M2 polarization in macrophages. IRF4 is a critical transcription factor of M2 polarization. A study has identified that JMJD3 induces H3K27 demethylation (H3K27me3) at the transcription factor IRF4 locus, which facilities M2 gene expression.72 Recently, IL-4-activated STAT6 was found to act as a transcriptional repressor in an HDAC3-dependent manner.73
Figure 4.
Epigenetic regulators of acetylation and methylation during macrophage polarization. Emerging evidence showed the histone acetylation and methylation enzymes modifying the various sites of histone tails to drive macrophage polarization.
Interestingly, histone modification has the standard and distinct macrophage enhancer repertoires. As an example, Spi1 enhancer, controlling the expression of PU.1, is marked by H3K4me2 and H3K27ac in all macrophage populations. H3K4me2 in all macrophage populations marks the RA-inducible Rarb gene, but high H3K27ac is only observed in large peritoneal macrophages and spleen macrophages, indicating a specific role of local RA in enhancer activation.74 Together, macrophages gain the ability to shift rapidly between cellular programs with epigenetic changes, suggesting epigenetic modification is a primary mechanism to affect phenotype plasticity.75,76
Critical transcription factors in macrophage polarization
Transcription factors and signals in M1 polarization
As mentioned above, epigenetic regulation of macrophages is an essential event for homeostasis and disease development. Consequently, as the collaborator of epigenetic regulation, transcription factors govern macrophage phenotypes and functions; they decide the transcriptomic profile to adapt to the homeostasis or disease conditions.
Macrophages polarize into distinct phenotypes in response to complex environmental cues; therefore, the in vitro polarization systems are too challenging to match fully the in vivo macrophage subsets. Nevertheless, as extremely polarized cells, M1 and M2 classify is still helpful in understanding the pro-inflammatory and anti-inflammatory status in homeostasis and diseases. The current studies of the M1 polarization mechanism are majorly based on classical activation (LPS+IFN-γ), which induces typical M1 phenotypes, including many well-known cytokines including IL1β, NOS2, IL-12, IL-6, and TNF-alpha (Il1β, nos2, Il12, Il6, and Tnf). The classical activation also triggers chemokines and chemokine receptors such as Ccl3, Cxcl10, Cxcl11, Ccl25, Cx3cr1, and Ccr7.
The LPS/IFN-γ triggers M1 polarization by activating several pathways: (1) NFκB pathway. LPS-TLR4-MYD88 is a classical cascade to active NF-κB signal. Based on studies using MyD88-deficient macrophages, proinflammatory cytokine expression was shown depending on the MyD88 pathway. Interestingly, the MyD88-independent pathway mediates the induction of type I IFNs and IFN-inducible genes. The study has shown that LPS/TLR4 is targeting STAT1-α/β in a MyD88 independent fashion. Both MyD88-dependent and MyD88-independent pathways activate IκB kinase (IKK) to phosphorylate IκB and liberate NFκB. Importantly, LPS indorses the feed-forward signaling of NF-κB by prompting autocrine cytokines, IL-1β and TNF-α, to sustain NF-κB activity.77 (2) JAK/STAT signal. STAT1 is an important transcription factor of M1 polarization induced by IFN-γ. IFN-γ signal induces JAK 1/2-mediated phosphorylation and dimerization of STAT1, which binds to the promoter of M1 signature genes [e.g., NOS2, IL-12, and class II transactivator (CIITA)]. IFN-γ-signal represses M2 genes were also reported.78 It has been demonstrated that activation of STAT1 promotes an inflammatory response in various diseases, such as atherosclerosis and inflammatory bowel disease.79,80 (3) IFN Regulatory Factors (IRF). IRF5, a critical transcription factor, is upregulated in M1 macrophages; it is involved in IL-12, IL-23, and TNF induction81 and Th1 and Th17 responses. Moreover, Irf5 deficiency in myeloid cells prevents necrotizing enterocolitis by inhibiting M1 macrophage polarization.82 A recent study found YAP (Yes-associated protein) aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. As a critical transcription coactivator, YAP's function in macrophage polarization is crucial for IBD therapy.83
Transcription factors and signals in M2 polarization
The molecular determinants of macrophage heterogeneity, specifically the transcription factors that dictate different responses to different environmental signals in different tissues, are still in a puzzle. Alternative activation (IL-4 or IL-4/IL-13) induces transcription of typical M2 macrophage genes, such as mrc1, retnlα, fizz1, chi3l3, and ym1, as well as phagocytosis ability. Further, STAT6, IRF4, PPARr, KLF4, and JMJD3, etc., transcription factors are clarified to be critical to M2 cell signatures.84 (1) STAT6. As the critical transcription factor in IL-4 or IL-4/IL-13 polarization system, Stat6 actives the transcription of genes typical of M2 polarization. Besides, STAT6 is also involved in gene repression by recruiting histone deacetylase (HDAC) complexes and decreasing enhancer accessibility. Asrar B Malik reported that activation of STAT6-mediated Gas6 expression during macrophage phenotype transition resulting in efferocytosis of PMNs plays a crucial role in resolving inflammatory lung injury.85 Stat6 interacts with Trim24 to modulates M2 phenotype and function, suggesting Stat6 acetylation is an essential negative regulatory mechanism that restricts M2 polarization. (2) PPAR-γ. PPAR-γ is a nuclear factor, it is constitutively expressed at a low level in macrophages, but its expression can be induced by IL-4 and IL-13, which indicates its potential role in M2 polarization in the setting of Th2 cell responses.86 Interestingly, a recent study has shown STAT6 as a cofactor involving PPARγ mediated gene regulation in vitro. Therefore, crosstalk between the IL-4–STAT6 axis and PPARγ might coordinate control the M2 phenotype. In PPAR-γ KO mice, macrophages are resistant to M2 polarization; the mice developed insulin resistance when challenged with a high-fat diet,69 and finally, they acquire an obesity phenotype. This metabolic phenotype is attributed to altered glucose metabolism in muscle and inflammation in adipose tissue, demonstrating a protective role for alternatively activated macrophages in obesity-associated metabolic disease.87,88 Interestingly, a recent study found that lung tumor cell-derived exosomes promote M2 macrophage polarization, which adds a novel regulation mechanism.89
Cutting-edge technologies for tissue macrophage study
Lineage tracing and genetic-fate mapping
According to the updated paradigm, tissue macrophages in most adult organs have two origins (1) embryonic-derived macrophages (residential- MΦ) that self-renew in situ, and (2) the circulating monocyte-progenitors-derived macrophages. Recent studies have shown that, in certain conditions, infiltrating monocyte-derived MΦ can supplement embryonic residential MΦ but remain divergent. However, the phenotypic and functional heterogeneity of tissue-residential macrophage versus their supplemented monocyte-derived MΦ counterparts in each organ system has not been recognized.17 A study has addressed this gap; the researchers generated a novel dual-reporter lineage tracing model to define residential-MΦ vs. monocyte-derived MΦ in vivo. This dual reporter approach uses a non-genotoxic method to deplete HSC in the Runx1Cre/eGFP mouse, allowing transplant LT-HSCs to express another color-mRFP1. Combining with high-dimensional flow cytometric analyses and single-cell mRNA sequencing, peritoneal MΦ showed further heterogeneity across lineages, supporting the thought that infiltrating monocyte-derived MΦ displays phenotypic difference from residential-macrophage potential functional impact.17 A similar fate-mapping approach has identified self-maintaining gut macrophages that are not replaced by monocytes.90 Therefore, lineage tracing is an essential tool for understanding macrophages' heterogeneity with different developmental origins in vivo,91 this innovative model incorporating lineage-specific fluorescent reporters in radiation-naïve mice showed a powerful potential in this field.
Flow-cytometry and CyTOF
In an immune cell study, flow cytometry is an extensively used approach to profile surface markers and phenotypes; however, the number of parameters limits its application in advanced analysis. Recently, the introduction of mass cytometry (CyTOF) has facilitated the high dimensional and unbiased investigation of the immune system, allowing real-time examination of many markers, which is vital for identifying phenotype from limited sample size and profound interrogation of immune responses. These two technologies are also essential for single-cell profiling in scRNA-seq. A new study using a combination of scRNA-seq and CyTOF in freshly isolated human microglia found remarkable differences in transcriptional states across different anatomical localization,92 indicating the spatial variety of human microglia. Interestingly, among the defined microglia clusters, CD68 and HLA-DR are highly expressed in white matter microglia compared with gray matter microglia, a similar finding of observations in mice.93 A group recently provided another comprehensive analysis of human microglia with CyTOF-based technology.94 This study illustrated the regional heterogeneity of microglia in the human brain by single-cell phenotyping of barcoded microglia from five brain regions. Intriguingly, the white matter zone and CNS region showed a dramatic difference in microglial cells95 on HLA-DR, CD68, CD11c, CD64, and the proliferation markers cyclin A and cyclin B1 expression, suggesting distinct functional features of microglia in a different location.94 In general, the application of flow-cytometry and CyTOF integrating other cutting-edge technologies such as scRNA-seq further deepens our understanding of the disease's immune cells phenotype and functions.
Single-cell RNA-sequencing
As the most heterogeneous population, macrophages differentiation, priming, and activation are dynamic changing patterns. Therefore, tracing the subsets change needs a high-dimensional and unbiased technology. The primary technologies identifying new cell subsets relied on cell surface markers staining and flow cytometry technology with a long history. Though these approaches were powerful, they required prior presumption of knowledge to uncover different immune cell types. Nevertheless, now an alternative workflow, single-cell sequencing (scRNA-seq), has revealed its advantage in the macrophage study. Due to the development of next-generation sequencing (NGS), the cost of sequencing is much decreased.
Meanwhile, the development of low-input RNA-seq protocols paved the way for further optimization down to the single-cell level, culminating in an outburst of new scRNA-seq platforms. Single-cell sequencing allowed cell sequencing without prior information of genes and proteins of interest and grouping of cells based on their transcriptional signatures. Promisingly, it allowed for the profiling of various rare cell populations using only 1 ng of RNA isolated from 100–1000 immune cells. When choosing the sequencing methods, there are many considerations, including the current single-cell profiling technologies, the experimental scale, the cost, the quality, and the sensitivity for data analysis. For instance, to sequence high breadth (a large number of cells) at low coverage is suitable for the studies aiming to identify cell clusters; on the opposite, studies aiming to distinguish stochastic variation in individual genes require a high sequencing depth.96
ScRNA-seq has the following application in immune cell study. (1) Characterizing cell subsets and cell states. Using scRNA-seq, Bjorklund et al. found four distinct ILC clusters with transcriptional profiles corresponding to previously characterized ILC1, ILC2, ILC3, and NK cell populations (based on surface marker expression).97 Another example, Gaublomme et al. used scRNA-seq to identify the T helper 17 (TH17) cell states that drive the pathogenesis of experimental autoimmune encephalomyelitis.98 (2) Describing the heterogeneity of a population. scRNA-seq can identify the diversity of macrophage subsets and adding a new dimension to understanding the macrophage dissection. For example, a group identified that by using unique transcriptional profiles, they found at least two unique tissue-resident interstitial macrophages in the steady-state lung that could be distinguished by and spatially localized to the interstitium of the bronchovascular bundles, but not alveolar walls.99 (3) Dissecting cell fate branch points. Trajectory analysis of scRNA-seq data can trace the dynamic cell differentiation track; in combination with the genetic fate-mapping, it can dissect the cell fate at branch points. Overall, the above studies show the power of scRNA-seq in reconstructing lineage trajectories and branching points and identifying previously unknown transcription factors that control transitions from one cellular state to the next in immune system development. Therefore, scRNA-seq is influential on characterizing homogeneous immune cell populations in health and disease, discovering the variation in stochastic gene expression that drives immunological responses, and tracing the cell fate branch points.
The macrophage is a well-target cell population for the scRNA-seq application. So far, many studies have uncovered heterogeneous transcriptional signatures in macrophages by using this technology. AMs have been the main research focus of lung Mφs, while IMs have been relatively overlooked in the lung. This was due partly to the abundance, accessibility, and ease of isolation of AMs compared to IMs, the latter that is much rarer and required tissue digestion and careful identification. However, with advances in high parameter flow cytometry and single-cell sequencing, IMs' importance has been recently re-evaluated. For example, by using single-cell RNA sequencing of both immune and non-immune cells in the developing lung, Cohen et al mapped candidate cell-cell interactions during alveolar macrophage development. This revealed potential cross-talk between epithelial cells, ILC2s, basophils, and the developing macrophages, validated both in vitro and in vivo.100 Lately, single-cell RNA-sequencing has shed new light on understanding the heterogeneity of human hepatic macrophages. In the human liver, hepatic macrophages consist of CD68+MARCO+ KCs, CD68+MARCO− macrophages, and CD14+ monocytes.101,102 CD68+MARCO+ KCs are characterized by enriched expression of genes involved in maintaining immune tolerance (e.g., VSIG4) and suppressing inflammation (e.g., CD163 and HMOX1). CD68+MARCO− macrophages have a similar transcriptional profile (e.g., C1QC, IL-18, S100A8/9) as recruited proinflammatory macrophages.101,102 However, both CD68+MARCO− macrophages and hepatic CD14+ monocytes show significantly weaker proinflammatory responses than circulating CD14+ monocytes.102
In summary, the insights from single-cell transcriptomics, advanced flow cytometry, lineage tracing methods greatly improved our understanding of the macrophage heterogeneous and their functions in homeostasis, injury, and inflammation resolution.
iPSC-derived primitive macrophages (iMacs)
It is well accepted that heterogeneous origins, tissue-specific factors, and microenvironmental elements determine the tissue macrophage populations and subpopulations, which have unique phenotypes and functions in different tissues. With the discovery that some tissue macrophages are not always replenished by circulating monocytes, even it was thought to be the case, but are instead long-lived self-maintaining cells that were seeded initially by early yolk sac (YS) or fetal liver during fetal development, the correspondingly study methods are also updated.
Tissue macrophages have been studied in vitro for many years, but they were hampered by the lack of access to enough appropriate cells for comprehensive in vitro at homeostasis. Therefore, bone marrow-derived macrophages (BMDM) or blood monocytes that have been cultured with specific cytokines and growth factors are widely used in previous and current work to study tissue-resident macrophages. In particular, human studies have relied heavily on the study of macrophages derived from blood monocyte cultures. However, this traditional approach was questioned recently. The tissue macrophages are generally not in a large population. The monocyte source is not readily available for human studies; moreover, the isolation process profoundly affects tissue-macrophage gene expression patterns, calling for caution in interpreting the results of studies relying on such methods.
With the need for a new approach to tissue-macrophage study, an innovative method has been considered, and emerging shreds of evidence have supported its potential as a promising platform. Induced pluripotent stem cells (iPSCs), showing the process of in vitro iMac differentiation, also closely mirrors the developmental pathways of the in utero YS program. Therefore, iPSC-derived primitive macrophage (iMacs) is a powerful tool in studying macrophages’ tissue-specific functions with the embryonic origin.103 Promisingly, the early phenotypic and transcriptomic analyses found that human iMacs shared common signature to fetal liver-derived monocytes than adult blood monocytes, indicated a more embryonic origin, engaging the iMacs closest to fetal liver monocytes, especially in terms of their expression of genes associated with tissue remodeling and angiogenesis. Importantly, analysis of mouse iMacs derived in a comparable way to the human iMacs similarly implied at the embryonic origin of these cells, with the iMacs showing remarkable transcriptomic similarity to macrophages isolated from the yolk sac. Recent evidence supports the potential of iMac in the tissue-macrophage study. For example, a research group generated macrophages from induced pluripotent stem cells (iPSCs) of an infantile-onset IBD patient lacking a functional IL10RB gene. These IL-10RB−/− Mϕs exhibited a striking defect in their ability to kill Salmonella enterica serovar Typhimurium. They further identify a regulatory interaction between IL-10 and PGE2, dysregulation of which may drive aberrant Mϕ activation and impaired host defense contributing to IBD pathogenesis.104 This study suggests that the iPSC-derived macrophage is a suitable model of macrophage ontogeny and heterogeneity for exploring cellular mechanisms and disease pathogenesis.
The iMac system has also been used to validate some critical transcription factors in the human study, such as PPARγ, and BACH2 for alveolar macrophages, ID3 for Kupffer cells. In the combination of the CRISPR–Cas9 gene-editing technology, as well as the bulk and single-cell transcriptomic analysis and the development of advanced tissue organoids, it is becoming promising to understand the mechanism involved in tissue-resident macrophage differentiation and reprogramming and how their dysregulation can result in both tissue and immune pathologies in humans. Taken together, studies formerly restricted to bone marrow cells and transgenic mouse models could now use the iMac system for the exploration of tissue-residential macrophages.
Nanoparticles in the macrophage study
With the nanomaterial's development, nanomedicine has become a promising field for solving clinical problems. Targeting macrophages by nanoparticles for a therapeutic reagent delivery has advanced our understanding of macrophage regulation mechanism and the potential for therapeutic purpose. There are several benefits of using nanoparticles in macrophage study: (1) Macrophage is a unique immune cell population, exists in many tissues with important biological functions. (2) Macrophages express extensive scavenger receptors and can uptake nanoparticles. (3) Properties of macrophages, including the heterogeneity, plasticity, and phagocytotic ability, make macrophages a potential target for nanomaterials delivery. The status of the nanoparticles in the macrophages study has been updated—first, the Spherical-Nano-particles-miRNA approach. In the study, researchers designed gold-core spherical nanoparticles, which coated with miR99bs. This miRNA can inhibit the MFG-E8 gene expression at the post-transcriptional level. In combination with in vivo study, they delivered spherical-nanoparticles-miRNA to the gut macrophages and illustrated a microRNA regulation mechanism in MFG-E8 protein and enterocyte migration.105 The nanoparticle-siRNA approach was also used in identifying mechanisms in skin disease.106 Second, a macrophage-like nanoparticle sponge. Cell membrane-coated nanoparticles have recently developed as a biomimetic nanomedicine platform, enabling a wide range of bio detoxification applications. These nanoparticles, acting as macrophage decoys, bind and neutralize endotoxins and proinflammatory cytokines, therefore, can inhibit inflammatory disease development. A study has illustrated macrophage-like nanoparticles concurrently absorbing endotoxins and proinflammatory cytokines for sepsis management.107 So, this strategy shows promise for improving patient outcomes, potentially shifting the current paradigm of sepsis management, and third, reprogramming of macrophages by nanoparticles. Macrophage subtypes are dissimilar in different tissues, especially for tumors. Tumor-associated macrophages (TAMs) usually express an M2 phenotype, enabling them to perform immunosuppressive and tumor-promoting functions. Switching these TAMs toward an M1 phenotype could enhance their antitumor immunity; however, it has been a challenge to achieve this reprogramming effect in vivo. Stephan et al describe a targeted nanocarrier that can deliver in vitro-transcribed mRNA encoding M1-polarizing transcription factors to reprogram TAMs without causing systemic toxicity.108 Ahmed MS et al. used a similar strategy in their study; they illustrated that TLR7/8-agonist-loaded nanoparticles promote tumor-associated macrophages' polarization to enhance cancer immunotherapy.109 Therefore, reprogramming of TAMs, also called re-education of TAMs, is currently the most striking strategy for tumor therapy and can reverse the protumor phenotype into the antitumor phenotype.
Collectively, the technologies stated here have advanced our understanding of the tissue-macrophage origins, differentiation and activation, mechanisms, and the role in the pathogenesis of diseases and provided the possibility for targeting macrophages as a therapeutic agent.
Summary and perspective
In this review, we first summarized the origins of tissue macrophages and their characters in different tissues. We provide insights into the diverse developmental origins of tissue macrophage, and this fundamental composition of mixed origins decides their need for renewal during homeostasis and disease. Second, we claim the classical molecular mechanisms of tissue macrophage differentiation and activation. From the environmental cues to epigenetic regulation and transcriptome change, any key element in this cascade will potentially modify macrophage phenotypes and functions in tissue, which is essential for translational medicine. Finally, we reviewed the up-to-date, cutting-edge technologies in the tissue macrophage study. These technologies deepen our understanding of macrophage heterogeneity and help us use these novel findings in therapeutic target identifying.
In the future, targeting tissue macrophage will be a promising therapeutic approach for inflammatory diseases. Macrophages represent a crucial component of acute inflammatory infiltration and are responsible for tissue injury and wound healing. Therefore, identifying the molecular mechanism of tissue macrophage differentiation, priming, activation, and repression are essential for clinical purposes. The present open questions and focuses are as follows: (1) Besides the gut, whether microbiota and metabolites will shape other tissue macrophage phenotypes and functions. (2) Whether the macrophage subsets are switchable and having the potential to be therapeutic targets. (3) The role of macrophages in immune tolerance and trained immunity. The multi-hits of bacteria or stress signals are very common in the pathogenesis of chronic inflammatory diseases, which challenge the host defense a second time and decide the disease's outcomes. However, the molecular- and immuno-mechanism about this concept is mostly unknown. Experimental evidence is needed to provide insights and understanding, and it will finally help us use tissue macrophages as a therapeutic tool.
Author contributions
All the authors made substantial contributions to the conception and writing of the article. All authors contributed to the initial draft and will contribute to the revisions. All authors agree to be accountable for all aspects of the work, including ensuring that the content is accurate.
Conflict of interests
The authors declare that there is no conflict of interest.
Funding
Xiao Wang is funded by US National Institutes of Health (NIH)-NIAID R21AI151943.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Abbreviations
- Mϕ
Macrophage
- YS
Yolk sac
- BMDM
Bone marrow-derived macrophage
- AMs
Alveolar Mφs
- IMs
Interstitial Mφs
- KC
Kupffer cells
- MoMφs
Monocyte-derived macrophages
- LCMs
Liver capsular macrophages
- PPARγ
Peroxisome proliferator receptor γ
- CCR2
CC-chemokine receptor 2
- LY6C
Lymphocyte antigen 6C
- MHCII
Major histocompatibility complex class II
- CX3CR1
CX3C-chemokine receptor 1
- TIM4
T cell immunoglobulin mucin receptor 4
References
- 1.Guilliams M., Thierry G.R., Bonnardel J., Bajenoff M. Establishment and maintenance of the macrophage niche. Immunity. 2020;52(3):434–451. doi: 10.1016/j.immuni.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Davies L.C., Jenkins S.J., Allen J.E., Taylor P.R. Tissue-resident macrophages. Nat Immunol. 2013;14(10):986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilliams M., Scott C.L. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol. 2017;17(7):451–460. doi: 10.1038/nri.2017.42. [DOI] [PubMed] [Google Scholar]
- 4.Ginhoux F., Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44(3):439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 5.Ginhoux F., Greter M., Leboeuf M., et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epelman S., Lavine K.J., Randolph G.J. Origin and functions of tissue macrophages. Immunity. 2014;41(1):21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez Perdiguero E., Klapproth K., Schulz C., et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518(7540):547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haldar M., Murphy K.M. Origin, development, and homeostasis of tissue-resident macrophages. Immunol Rev. 2014;262(1):25–35. doi: 10.1111/imr.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locati M., Curtale G., Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perdiguero E.G., Geissmann F. The development and maintenance of resident macrophages. Nat Immunol. 2016;17(1):2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapouri-Moghaddam A., Mohammadian S., Vazini H., et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233(9):6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 12.Bain C.C., Schridde A. Origin, differentiation, and function of intestinal macrophages. Front Immunol. 2018;9:2733. doi: 10.3389/fimmu.2018.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Barres B.A. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018;18(4):225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 14.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J. Microglia in neurodegeneration. Nat Neurosci. 2018;21(10):1359–1369. doi: 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan S.Y., Krasnow M.A. Developmental origin of lung macrophage diversity. Development. 2016;143(8):1318–1327. doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misharin A.V., Morales-Nebreda L., Reyfman P.A., et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214(8):2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yona S., Kim K.W., Wolf Y., et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38(1):79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bharat A., Bhorade S.M., Morales-Nebreda L., et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol. 2016;54(1):147–149. doi: 10.1165/rcmb.2015-0147LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaitin D.A., Adlung L., Thaiss C.A., et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178(3):686–698. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misharin A.V., Morales-Nebreda L., Mutlu G.M., Budinger G.R., Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49(4):503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyfman P.A., Walter J.M., Joshi N., et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(12):1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S.S., Lv X.X., Liu C., et al. Targeting degradation of the transcription factor C/EBPβ reduces lung fibrosis by restoring activity of the ubiquitin-editing enzyme A20 in macrophages. Immunity. 2019;51(3):522–534. doi: 10.1016/j.immuni.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura A., Ebina-Shibuya R., Itoh-Nakadai A., et al. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J Exp Med. 2013;210(11):2191–2204. doi: 10.1084/jem.20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi T., Denney L., An H., Ho L.P., Zheng Y. Alveolar and lung interstitial macrophages: definitions, functions, and roles in lung fibrosis. J Leukoc Biol. 2021;110(1):107–114. doi: 10.1002/JLB.3RU0720-418R. [DOI] [PubMed] [Google Scholar]
- 25.Westphalen K., Gusarova G.A., Islam M.N., et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506(7489):503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neupane A.S., Willson M., Chojnacki A.K., et al. Patrolling alveolar macrophages conceal bacteria from the immune system to maintain homeostasis. Cell. 2020;183(1):110–125. doi: 10.1016/j.cell.2020.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Schyns J., Bai Q., Ruscitti C., et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat Commun. 2019;10(1):3964. doi: 10.1038/s41467-019-11843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krenkel O., Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17(5):306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 29.Zigmond E., Samia-Grinberg S., Pasmanik-Chor M., et al. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193(1):344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 30.Scott C.L., Zheng F., De Baetselier P., et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. doi: 10.1038/ncomms10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nascimento M., Huang S.C., Smith A., et al. Ly6Chi monocyte recruitment is responsible for Th2 associated host-protective macrophage accumulation in liver inflammation due to schistosomiasis. PLoS Pathog. 2014;10(8):e1004282. doi: 10.1371/journal.ppat.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stutchfield B.M., Antoine D.J., Mackinnon A.C., et al. CSF1 restores innate immunity after liver injury in mice and serum levels indicate outcomes of patients with acute liver failure. Gastroenterology. 2015;149(7):1896–1909. doi: 10.1053/j.gastro.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You Q., Cheng L., Kedl R.M., Ju C. Mechanism of T cell tolerance induction by murine hepatic Kupffer cells. Hepatology. 2008;48(3):978–990. doi: 10.1002/hep.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heymann F., Peusquens J., Ludwig-Portugall I., et al. Liver inflammation abrogates immunological tolerance induced by Kupffer cells. Hepatology. 2015;62(1):279–291. doi: 10.1002/hep.27793. [DOI] [PubMed] [Google Scholar]
- 35.Wen Y., Lambrecht J., Ju C., Tacke F. Hepatic macrophages in liver homeostasis and diseases-diversity, plasticity and therapeutic opportunities. Cell Mol Immunol. 2021;18(1):45–56. doi: 10.1038/s41423-020-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beattie L., Sawtell A., Mann J., et al. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J Hepatol. 2016;65(4):758–768. doi: 10.1016/j.jhep.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuinness P.H., Painter D., Davies S., McCaughan G.W. Increases in intrahepatic CD68 positive cells, MAC387 positive cells, and proinflammatory cytokines (particularly interleukin 18) in chronic hepatitis C infection. Gut. 2000;46(2):260–269. doi: 10.1136/gut.46.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim A., Wu X., Allende D.S., Nagy L.E. Gene deconvolution reveals aberrant liver regeneration and immune cell infiltration in alcohol-associated hepatitis. Hepatology. 2021;74(2):987–1002. doi: 10.1002/hep.31759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bian Z., Gong Y., Huang T., et al. Deciphering human macrophage development at single-cell resolution. Nature. 2020;582(7813):571–576. doi: 10.1038/s41586-020-2316-7. [DOI] [PubMed] [Google Scholar]
- 40.Garrett W.S., Gordon J.I., Glimcher L.H. Homeostasis and inflammation in the intestine. Cell. 2010;140(6):859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Schepper S., Verheijden S., Aguilera-Lizarraga J., et al. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell. 2018;175(2):400–415. doi: 10.1016/j.cell.2018.07.048. [DOI] [PubMed] [Google Scholar]
- 42.Na Y.R., Stakenborg M., Seok S.H., Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat Rev Gastroenterol Hepatol. 2019;16(9):531–543. doi: 10.1038/s41575-019-0172-4. [DOI] [PubMed] [Google Scholar]
- 43.Shaw T.N., Houston S.A., Wemyss K., et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med. 2018;215(6):1507–1518. doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sehgal A., Donaldson D.S., Pridans C., Sauter K.A., Hume D.A., Mabbott N.A. The role of CSF1R-dependent macrophages in control of the intestinal stem-cell niche. Nat Commun. 2018;9(1):1272. doi: 10.1038/s41467-018-03638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadis U., Wahl B., Schulz O., et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34(2):237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Shaw M.H., Kamada N., Kim Y.G., Núñez G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209(2):251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortha A., Chudnovskiy A., Hashimoto D., et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343(6178):1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulthess J., Pandey S., Capitani M., et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50(2):432–445. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang B., Alvarado L.J., Kim T., et al. Commensal microbiota drive the functional diversification of colon macrophages. Mucosal Immunol. 2020;13(2):216–229. doi: 10.1038/s41385-019-0228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Li X., Luo Z., et al. ECM1 is an essential factor for the determination of M1 macrophage polarization in IBD in response to LPS stimulation. Proc Natl Acad Sci U S A. 2020;117(6):3083–3092. doi: 10.1073/pnas.1912774117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koelink P.J., Bloemendaal F.M., Li B., et al. Anti-TNF therapy in IBD exerts its therapeutic effect through macrophage IL-10 signalling. Gut. 2020;69(6):1053–1063. doi: 10.1136/gutjnl-2019-318264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray P.J. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 53.Ginhoux F., Schultze J.L., Murray P.J., Ochando J., Biswas S.K. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17(1):34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 54.Phan A.T., Goldrath A.W., Glass C.K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46(5):714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saeed S., Quintin J., Kerstens H.H., et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345(6204):1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su S., Zhao Q., He C., et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun. 2015;6:8523. doi: 10.1038/ncomms9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minutti C.M., Jackson-Jones L.H., García-Fojeda B., et al. Local amplifiers of IL-4Rα-mediated macrophage activation promote repair in lung and liver. Science. 2017;356(6342):1076–1080. doi: 10.1126/science.aaj2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masud S., Prajsnar T.K., Torraca V., et al. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy. 2019;15(5):796–812. doi: 10.1080/15548627.2019.1569297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray P.J., Allen J.E., Biswas S.K., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banerjee S., Cui H., Xie N., et al. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem. 2013;288(49):35428–35436. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jablonski K.A., Gaudet A.D., Amici S.A., Popovich P.G., Guerau-de-Arellano M. Control of the inflammatory macrophage transcriptional signature by miR-155. PLoS One. 2016;11(7):e0159724. doi: 10.1371/journal.pone.0159724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D., Duan M., Feng Y., Geng L., Li X., Zhang W. MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA. Mol Immunol. 2016;77:205–212. doi: 10.1016/j.molimm.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Zhang D., Tang Z., Huang H., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lavin Y., Winter D., Blecher-Gonen R., et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai W., Dai X., Chen J., et al. STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight. 2019;4(20):e131355. doi: 10.1172/jci.insight.131355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dorrington M.G., Fraser I.D.C. NF-κB signaling in macrophages: dynamics, crosstalk, and signal integration. Front Immunol. 2019;10:705. doi: 10.3389/fimmu.2019.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447(7148):1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saleiro D., Platanias L.C. Intersection of mTOR and STAT signaling in immunity. Trends Immunol. 2015;36(1):21–29. doi: 10.1016/j.it.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daskalaki M.G., Tsatsanis C., Kampranis S.C. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J Cell Physiol. 2018;233(9):6495–6507. doi: 10.1002/jcp.26497. [DOI] [PubMed] [Google Scholar]
- 72.Satoh T., Takeuchi O., Vandenbon A., et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 73.Czimmerer Z., Daniel B., Horvath A., et al. The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity. 2018;48(1):75–90. doi: 10.1016/j.immuni.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gosselin D., Link V.M., Romanoski C.E., et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159(6):1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li T., Garcia-Gomez A., Morante-Palacios O., et al. SIRT1/2 orchestrate acquisition of DNA methylation and loss of histone H3 activating marks to prevent premature activation of inflammatory genes in macrophages. Nucleic Acids Res. 2020;48(2):665–681. doi: 10.1093/nar/gkz1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Placek K., Schultze J.L., Aschenbrenner A.C. Epigenetic reprogramming of immune cells in injury, repair, and resolution. J Clin Invest. 2019;129(8):2994–3005. doi: 10.1172/JCI124619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toshchakov V., Jones B.W., Perera P.Y., et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3(4):392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 78.Kang K., Park S.H., Chen J., et al. Interferon-γ represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity. 2017;47(2):235–250. doi: 10.1016/j.immuni.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivashkiv L.B. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018;18(9):545–558. doi: 10.1038/s41577-018-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stark G.R., Darnell J.E. Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krausgruber T., Blazek K., Smallie T., et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12(3):231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 82.Wei J., Tang D., Lu C., et al. Irf5 deficiency in myeloid cells prevents necrotizing enterocolitis by inhibiting M1 macrophage polarization. Mucosal Immunol. 2019;12(4):888–896. doi: 10.1038/s41385-019-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou X., Li W., Wang S., et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. 2019;27(4):1176–1189. doi: 10.1016/j.celrep.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 84.Liao X., Sharma N., Kapadia F., et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121(7):2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nepal S., Tiruppathi C., Tsukasaki Y., et al. STAT6 induces expression of Gas6 in macrophages to clear apoptotic neutrophils and resolve inflammation. Proc Natl Acad Sci U S A. 2019;116(33):16513–16518. doi: 10.1073/pnas.1821601116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bouhlel M.A., Derudas B., Rigamonti E., et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 87.Szanto A., Balint B.L., Nagy Z.S., et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33(5):699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palmer J.G. Liability considerations presented by human gene therapy. Hum Gene Ther. 1991;2(3):235–242. doi: 10.1089/hum.1991.2.3-235. [DOI] [PubMed] [Google Scholar]
- 89.Pritchard A., Tousif S., Wang Y., et al. Lung tumor cell-derived exosomes promote M2 macrophage polarization. Cells. 2020;9(5):1303. doi: 10.3390/cells9051303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vannella K.M., Wynn T.A. Mechanisms of organ injury and repair by macrophages. Annu Rev Physiol. 2017;79:593–617. doi: 10.1146/annurev-physiol-022516-034356. [DOI] [PubMed] [Google Scholar]
- 91.Culemann S., Grüneboom A., Nicolás-Ávila J.Á., et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 2019;572(7771):670–675. doi: 10.1038/s41586-019-1471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sankowski R., Böttcher C., Masuda T., et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci. 2019;22(12):2098–2110. doi: 10.1038/s41593-019-0532-y. [DOI] [PubMed] [Google Scholar]
- 93.Goldmann T., Zeller N., Raasch J., et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34(12):1612–1629. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Böttcher C., Schlickeiser S., Sneeboer M.A.M., et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat Neurosci. 2019;22(1):78–90. doi: 10.1038/s41593-018-0290-2. [DOI] [PubMed] [Google Scholar]
- 95.Deczkowska A., Matcovitch-Natan O., Tsitsou-Kampeli A., et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat Commun. 2017;8(1):717. doi: 10.1038/s41467-017-00769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 97.Björklund Å.K., Forkel M., Picelli S., et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17(4):451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 98.Gaublomme J.T., Yosef N., Lee Y., et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163(6):1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ural B.B., Yeung S.T., Damani-Yokota P., et al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci Immunol. 2020;5(45):eaax8756. doi: 10.1126/sciimmunol.aax8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scott C.L., Guilliams M. Tissue Unit-ed: lung cells team up to drive alveolar macrophage development. Cell. 2018;175(4):898–900. doi: 10.1016/j.cell.2018.10.031. [DOI] [PubMed] [Google Scholar]
- 101.MacParland S.A., Liu J.C., Ma X.Z., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9(1):4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao J., Zhang S., Liu Y., et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020;6:22. doi: 10.1038/s41421-020-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takata K., Kozaki T., Lee C.Z.W., et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity. 2017;47(1):183–198. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 104.Mukhopadhyay S., Heinz E., Porreca I., et al. Loss of IL-10 signaling in macrophages limits bacterial killing driven by prostaglandin E2. J Exp Med. 2020;217(2):e20180649. doi: 10.1084/jem.20180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang X., Hao L., Bu H.F., et al. Spherical nucleic acid targeting microRNA-99b enhances intestinal MFG-E8 gene expression and restores enterocyte migration in lipopolysaccharide-induced septic mice. Sci Rep. 2016;6:31687. doi: 10.1038/srep31687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng D., Giljohann D.A., Chen D.L., et al. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci U S A. 2012;109(30):11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thamphiwatana S., Angsantikul P., Escajadillo T., et al. Macrophage-like nanoparticles concurrently absorbing endotoxins and proinflammatory cytokines for sepsis management. Proc Natl Acad Sci U S A. 2017;114(43):11488–11493. doi: 10.1073/pnas.1714267114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang F., Parayath N.N., Ene C.I., et al. Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nat Commun. 2019;10(1):3974. doi: 10.1038/s41467-019-11911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodell C.B., Arlauckas S.P., Cuccarese M.F., et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng. 2018;2(8):578–588. doi: 10.1038/s41551-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]