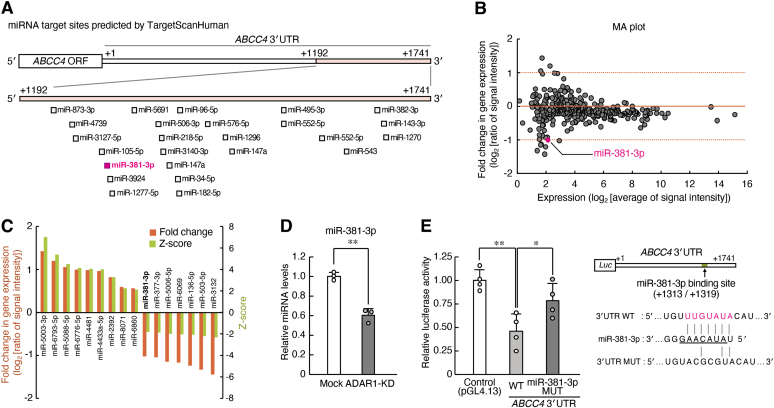
Figure 2.
ADAR1 regulates the translation of ABCC4 mRNA into MRP4 protein through mediating miR-381-3p expression.A, schematic diagram showing the putative target sites of miRNAs in ABCC4 3′UTR from +1192 to +1741. The numbers indicate the distance in base pairs from the stop codon (+1). The miRNA target sites were predicted with TargetScanHuman (Release 7.2). B, MA plot of all miRNAs expressed in mock-transduced and ADAR1-KD RPTECs. Data were obtained from microarray analysis (accession number GSE192692). The criteria for upregulated miRNAs were set at a ratio of 2-fold or Z-score of 2.0; the criteria for downregulated miRNAs were set at a ratio of 0.5-fold or Z-score of −2.0. C, differentially expressed miRNAs between mock-transduced and ADAR1-KD RPTECs in the microarray analysis. D, the expression levels of miR-381-3p in mock-transduced and ADAR1-KD RPTECs. The expression levels were assessed by qRT-PCR and normalized to those of miR-191-5p, which was used as an endogenous control of miRNA and normalizer of RT and PCR steps (32). Values are the mean with SD (n = 3). ∗∗p < 0.01; significant difference between the two groups (t4 = 8.567, p = 0.001; unpaired t test, two sided). E, the luciferase activity in RPTECs transfected with wildtype (WT) or mutant (MUT) ABCC4 3′UTR::Luc. Schematic diagrams of luciferase reporter constructs containing WT or MUT (mutation in miR-381-3p target site) 3′UTR of the ABCC4 gene are shown in the right panel. The numbers in the right panel indicate the distance in base pairs from the stop codon (+1). Values are the mean with SD (n = 4). ∗∗p < 0.01, ∗p < 0.05; significant difference between the indicated groups (F2, 9 = 11.252, p = 0.004; ANOVA with Tukey–Kramer’s post hoc test).